Research Article - (2024) Volume 14, Issue 5
Radiogenomic Prediction of MGMT Promoter Methylation Status in Glioblastoma Using Multi-Modal MRI and EfficientNet
- Corresponding Author:
- Pavan Nathani
Department of Neuroscience, Centennial High School, Frisco, United States of America
E-mail: [email protected]
Received date: 09-Oct-2024, Manuscript No. NPY-24-149887; Editor assigned: 11-Oct-2024, PreQC No. NPY-24-149887 (PQ); Reviewed date: 25-Oct-2024, QC No.NPY-24-149887; Revised date: 01-Nov-2024, Manuscript No. NPY-24-149887 (R); Published date: 08-Nov-2024, DOI: 10.37532/1758-2008.2024.14(5).737
Abstract
Methylguanine DNA Methyltransferase (MGMT) promoter hypermethylation is a critical biomarker for predicting temozolomide sensitivity in aggressive Glioblastoma Multiforme (GBM), a disease with a poor prognosis. A Methylguanine DNA Methyltransferase status prediction without biopsy is mandatory for tailored therapy because MGMT patients have better prognosis. To this end, this paper proposes a radiogenomic model for predicting MGMT status in Glioblastoma patients based on multimodal magnetic resonance imaging (MRI) data and the EfficientNet deep learning architecture. Using sophisticated machine learning-based non-invasive methods of genetic prognosis may decrease risky biopsies. The MRI sequences in the Brain Tumor Segmentation (BraTS) 21 competition dataset include T1-weighted (T1w), T1-weighted contrast-enhanced (T1wCE), T2-weighted (T2w), and Fluid Attenuated Inversion Recovery (FLAIR) scans. Image preprocessing consisted of normalization, scaling, and Fourier transform-based data augmentation for cross-modal alignment. The EfficientNet-B0 model was initially pre-trained on ImageNet and then fine-tuned to the binary classification of methylated and unmethylated MGMT. The models were evaluated based on training and validation scores. The model’s ability to accurately identify MGMT methylation status was moderate, with a validation score score of 0.62393 at its best. Applying multimodal MRI data elevated feature extraction, proving deep learning models have radiogenomic predictive power. This study demonstrates that Automatic Live Control (ALC) and Deep Learning (DL) models can non-invasively assess MGMT promoter methylation status. That being said, such models may suffer from overfitting or generalization, requiring optimization; however, they are still a safer approach to biopsies and can enhance Glioblastoma treatment.
Keywords
Glioblastoma, Radiogenomics, MGMT promoter methylation, EfficientNet
Introduction
Regrettably, Glioblastoma Multiforme (GBM) is one of the most malignant and frequent primary brain tumors in adults. Ependymomas constitute 15-20 percent of intracranial tumors and 55 percent of posterior fossa malignancies. Although recent developments in surgery, radiotherapy, and chemotherapy have contributed to the management of GBM, over 80 percent of patients survive only 12- 15 months after diagnosis. The five-year survival rate of GBM is below 5 percent [1- 3]. MGMT gene methylation status influences the extent to which tumors become sensitive to chemotherapy, specifically Temozolomide (TMZ). The MGMT gene codes for a DNA repair enzyme, which repairs the TMZ damage. When this gene is silenced by methylation, tumor cells lose their ability to repair TMZ- induced damage, making them more vulnerable to treatment [4,5]. However, if cancers lack this methylation, they have a higher production of MGMT, thus making chemotherapy ineffective and have a worse prognosis. This promoter methylation is present in 35 to 75 percent of GBM patients and is a significant predictor of the TMZ response.
An invasive biopsy and molecular tests such as pyrosequencing have always decided MGMT status. Biopsies are accurate, but the behavior of the tumor and its response leads to complications such as infection, bleeding, and error sampling. Consequently recent studies have shifted towards using noninvasive technologies and better imaging, such as multimodal MRI, to predict MGMT status without performing a biopsy [6]. Recent work has highlighted the ability to build models based on imaging, radiogenomics, and non- invasive MGMT methylation prediction [7].
Many equate MRI characteristics, including diffusion-weighted or perfusion-weighted images, with molecular characteristics of a tumor in hopes of altering GBM diagnosis and monitoring [8,9]. However, efficacy regarding specific imaging methodologies and procedures used to validate the model has led to accuracies between 60 percent and 85 percent across case studies. Prior radiogenomic approaches that employ hand-built features, such as intensity and texture, are deficient in capturing data details, resulting in low accuracy.
Current research only employs basic machine learning methods such as support vector machines and random forest methods while leaving deep learning architectures unexploited. Enhanced models are required to improve the forecast’s precision and model transferability.
Our study proposes the utilization of EfficientNet, a state of the art deep learning model, to predict MGMT promoter methylation status from multimodal MRI data. EfficientNet scales depth, width, and resolution with a compound scaling method to balance model complexity and efficiency, making it highly suitable for medical picture analysis. Here, we construct a non-invasive predictive model based on T1-weighted (T1w), T1-weighted contrast-enhanced (T1wCE), T2-weighted (T2w), and Fluid Attenuated Inversion Recovery (FLAIR) MRI scans. Using these modalities based on EfficientNet, we aim to improve MGMT methylation predictions for proper GBM diagnosis and treatment [10].
Materials and Methods
Study design and data source
This retrospective, observational study uses multimodal MRI studies from the Radiological Society of North America Medical Image Computing and Computer Assisted Intervention (RSNA-MICCAI) Brain Tumor Radiogenomic Classification Challenge 2021. MRI examinations include T1w, T1wCE, T2w, and FLAIR scans with MGMT promoter methylation labels. De- identified images from many clinical facilities were provided [11]. Ethical approval was not required because this dataset was freely available and anonymized.
Image pre-processing and normalization
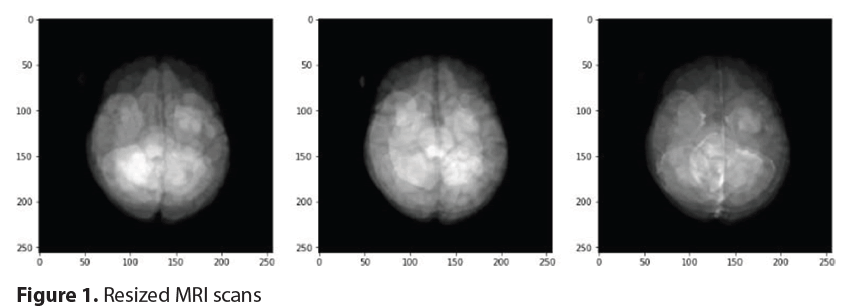
Figure 1 highlights how each image was resized to 256 × 256 pixels to standardize images across MRI modalities. Pixel intensity values were normalized in the range of 0-1, which significantly improved the model’s convergence during training. Regular rotations, horizontal flipping, and zooming helped prevent overfitting and improved the model’s ability to generalize data it would not have seen before [12].
Model architecture EfficientNet-B0
We used the EfficientNet-B0 architecture to predict MGMT promoter methylation status as it balances performance and computational efficiency well.
Unlike typical Convolutional Neural Networks (CNNs), EfficientNet-B0 employs compound scaling to uniformly scale network depth, width, and resolution for improved performance while minimizing computing costs. Given the sizeable nature of the MRI datasets , we prioritized efficiency.
EfficientNet-B0 was pre-trained on ImageNet and fine-tuned for binary classification of methylated or unmethylated MGMT utilizing BraTS 2021 MRI data.
The depth-wise separable convolutions maintained performance with with Mobile Inverted Bottleneck Convolution (MBConv) layers [13].
Training and cross-validation
We split the dataset into 80 percent training and 20 percent validation. To balance methylation and unmethylated instances across all folds, we used stratified k-fold cross-validation (k=5). The Adam optimizer, with a 0.001 learning rate and 16 batches, trained the model. Binary classification problems used binary cross- entropy as a loss function. Early stopping reduced overfitting by ending training when validation loss plateaued for over ten epochs [14].
Evaluation metrics
We used training and validation scores as our primary metrics to assess our model’s ability to distinguish between methylated and unmethylated cases.
Results
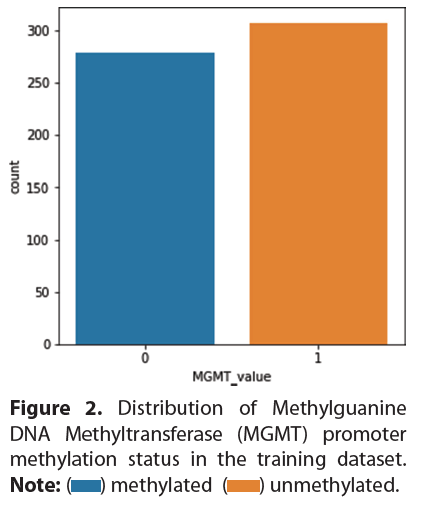
Distribution of MGMT promoter methylation status
The dataset we used for training contained a near even mix of methylated and unmethylated samples, as shown in Figure 2.
Sample MRI visualization
Figure 3 highlights examples of the different MRI modalities we worked with, including FLAIR, T1w, T1wCE, and T2w. Each type of scan captures unique details about the brain’s anatomy. For instance, FLAIR images are excellent for showing fluid accumulation while T1wCE images enhance contrast to define the tumor’s edges better. These variations are vital in identifying genetic traits like MGMT promoter methylation from the pictures. Combining different kinds of MRI data can develop a complete picture of the tumor and its characteristics.
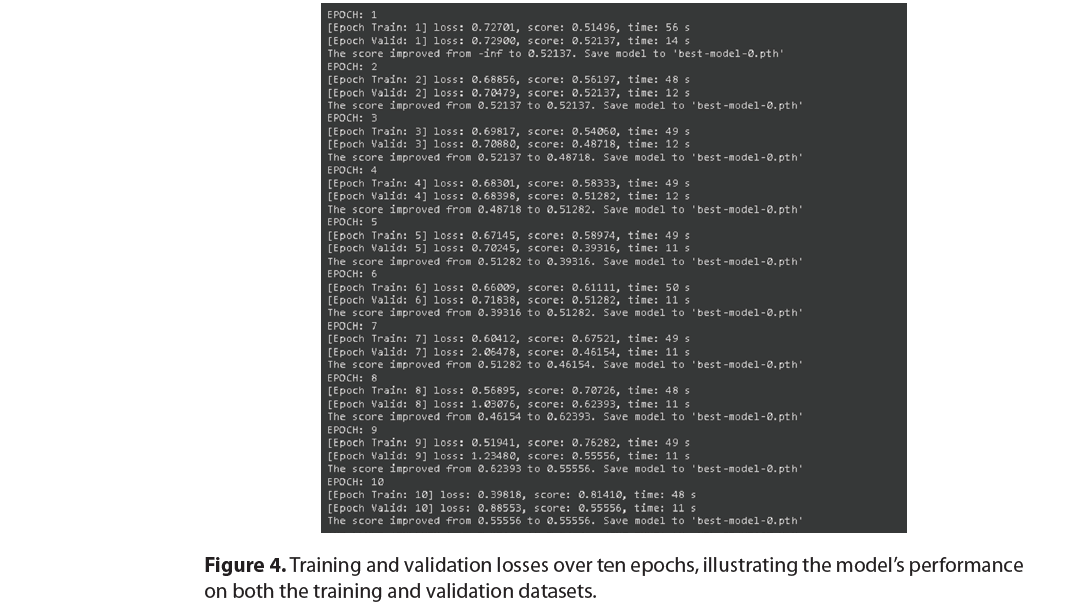
Training and validation loss
Figure 4 shows how the model’s loss evolved over ten training epochs, illustrating the model’s performance on both training and validation data. The training loss steadily decreased, which is a good sign that the model was learning. However, the validation loss did not follow the same pattern and fluctuated quite a bit, which could indicate overfitting. To combat this, we used early stopping.
Discussion
Interpretation of results
It is clear from the results of the training phase that the model’s performance gradually increased over time. However, the high accuracy and loss fluctuations in the validation phase demonstrate overfitting. The model improved its training and validation scores from epochs 1-10, which peaked at 0.8141 and 0.6239, respectively. However, in Epoch 5, the validation performance dropped to 0.3932, while improvements in training performance were recorded (Epoch5:0.5897). This indicates overfitting as the model internalized the training dataset quite well but could not universalize that learning to other spectrums of new datasets.
These results may be due to the varied nature of MRI imaging data and the challenges in achieving confident features for MGMT methylation prediction. Improvements in performance fluctuations could justify dropout techniques or other more assertive modes of early stopping. These changes may also enhance the model’s cross-dataset generalization and imaging method consistency.
Clinical implications
Deep learning algorithms like EfficientNet may help predict MGMT promoter methylation non-invasively in patients with GBM. As methylated tumors are more sensitive to TMZ, it is essential to determine MGMT methylation status in providing personalized chemotherapy. However, standard MGMT status tests require biopsies, carrying the risk of wound infection and sampling errors. As in our study, noninvasive MRI-based alternatives may improve treatment approach and prognosis by facilitating versatile treatment regimens based on the tumor`s methylation status, thus improving the chances of survival while preventing toxicity associated with treatment.
Limitations
The model performed well, but training and validation must be improved. Loss and accuracy fluctuations in validation metrics, especially after Epoch 7, indicate that the model overfit to the training set, suggesting an inability to generalize across complex heterogeneous imaging data. The small size and uniformity of the training dataset are likely the reasons for this overfitting. The RSNA-MICCAI dataset containing MRI data of GBM patients may not encompass the entire spectrum of patients with a clinical diagnosis of GBM, thus diminishing the model’s applicability to actual practice.
Conclusion
In conclusion, after the model was trained and validated, the predictions made by the EfficientNet model regarding MGMT promoter methylation status based on multi-modal MRI proved moderately accurate. The maximum validation score recorded during epoch eight was 0.62393, with low validation loss and some indication of overfitting. The training performance improved over time, reaching 0.81410 by the end of epoch 10. However, it must be emphasized that the validation scores indicate that additional steps are still needed to improve the model’s optimization and regularization This work demonstrates that radiogenomic deep learning models allow for critical gene status assessment in GBM, with the caveat that they must be improved before they can be used in the clinic.
Future Scope
Genomic and molecular information such as advanced molecular subtypes, Isocitrate Dehydrogenase (IDH1 or IDH2) mutations, or Epidermal Growth Factor Receptor (EGFR) amplification could be incorporated into the model to enhance its predictive power. Combining various biomarkers enhances patient outcomes and treatment response prediction. Model performance may be improved by utilizing more complex topologies, such as transformer networks that have the potential to capture long-range sequences in visual data. With federated learning, an institution can train models on data belonging to other institutions. This would help address data homogeneity, thus promoting model robustness in different clinical environments. Overfitting can be combatted and the general overview can be enhanced with dropout or L2 regularization.
Acknowledgements
The data for this study was obtained from the RSNA-MICCAI BraTS 2021, to which we are so thankful. We also thank opensource developers who have created tools and frameworks that made it possible to utilize the EfficientNet deep learning architecture.
Ethical Considerations
As the data utilized in this study was publicly available and fully anonymized, no Institutional Review Board (IRB) approval was required. Nevertheless, we ensured all data processing and analyses adhered to the highest data privacy standards and ethical research practices.
References
- Kumar A, Wei, J, Hao, X et al. Radiogenomics for predicting MGMT promoter methylation status in glioblastoma using CNN-based machine learning. IEEE Trans on Med Imag 39(7), 2176-2187 (2020). [Google Scholar]
- Yogananda CG, Shah BR, Nalawade SS, et al. MRI-based deep-learning method for determining glioma MGMT promoter methylation status. Am J Neuroradiol 42(5), 845-852 (2021).
- Tan M, Le Q. Efficientnet: Rethinking model scaling for convolutional neural networks. Int. Conf. Mach. Learn 6105-6114 (2019).
- Fang Q. The versatile attributes of MGMT: Its repair mechanism, crosstalk with other DNA repair pathways and its role in cancer. Cancers 16(2), 331 (2024).
- Moon WJ, Choi JW, Roh HG et al. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: The CT, diffusion tensor imaging and perfusion MR imaging. Neuroradiol 54, 555-563 (2012).
- Le Guillou Horn XM, Lecellier F, Giraud C, et al. From voxel to gene: A scoping review on MRI radiogenomics’ artificial intelligence predictions in adult gliomas and glioblastomas-the promise of virtual biopsy?. Biomedicines 12(9), 2156 (2024).
- Korfiatis P, Kline TL, Lachance DH, et al. Residual deep convolutional neural network predicts MGMT methylation status. J. Digit Imaging 30, 622-628 (2017).
- Doniselli FM, Pascuzzo R, Mazzi F, et al. Quality assessment of the MRI-radiomics studies for MGMT promoter methylation prediction in glioma: A systematic review and meta-analysis. Eur Radiol 1-4 (2024).
- Korfiatis P, Kline TL, Coufalova L, et al. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Medical phys 43(1), 2835-2844 (2016).
- Kaggle H, Capuozzo S, Gravina M, et al. A multimodal knowledge-based deep learning approach for MGMT promoter methylation identification. J. Imaging 8(12), 321 (2022).
- Feng X, Tustison NJ, Patel SH, et al. Brain tumor segmentation using an ensemble of 3d u-nets and overall survival prediction using radiomic features. Front. comput. Neurosci14:25 (2020).
- Khomidov M, Lee JH. The novel efficient net architecture-based system and algorithm to predict complex human emotions. Algorithms 17(7), 285 (2024).
- Saeed N, Hardan S, Abutalip K, et al. Is it possible to predict MGMT promoter methylation from brain tumor MRI scans using deep learning models?. Int Conf on Medical Imaging 1005-1018 (2022).
- Wang F, Jiang R, Zheng L, et al. 3d u-net based brain tumor segmentation and survival days prediction. Cham: Springer Int Publishing 131-141 (2019).
 ) methyalted; (
) methyalted; ( ) unmethylated.
) unmethylated.