Research Article - (2018) Volume 8, Issue 3
Pharmacophore Screening and Docking studies of AMPA Receptor Implicated in AlzheimerâÃâ¬Ãâ¢s disease with Some CNS Acting Phytocompounds from Selected Ayurvedic Medicinal Plants
- Corresponding Author:
- Preenon Bagchi
Padmashree Institute of Management and Sciences, Bangalore, India
Abstract
Neuro-degeneration including dementia in Alzheimer ’s disease (AD) is an global alarming problem. AD accounts for 60 to 80 percent of dementia cases. AMPA (alpha-amino-3-hydroxy- 5-methyl-4-isoxazole propionic acid) receptors are the main contributors of excitatory neurotransmission, mediating the fast, rapidly desensitizing excitation of many synapses, and are involved in the early response to glutamate in the synaptic space. The gene receptor AMPA responsible for AD is taken for this work. In the present study phyto-compounds from Ayurvedic Medicinal plants like Centella asiatica, Bacopa monnieri, Convolvulus pluricaulis, Mucuna pruriens, Ocimum sanctum, Tinospora cordifolia, Curcuma longa, Nardostachys jatamansi are among others used. The active components of the plants are taken and pharmacophore screening along with docking studies are performed against AMPA receptor in silico. The docking scores are noted for further in-vitro receptor-ligand binding assay studies. The selected phytocompounds were screened against AMPA receptor. Again, ADME (drug-like properties) was determined for the shortlisted ligands. Based on virtual screening, shortlisted ligands selected were Quercetin dihydrate and Asiatic acid.
Keywords
Alzheimer’s disease (AD), AMPA, Modeling, Ramachandran plot, Pharmacophore, docking, ADME
Introduction
Since ages various medicinal plants are part & parcel of major populations of India & other South East Asian countries. For the management of different neuro-degenerative diseases (Alzheimer’s, parkinsonism, obsessive compulsive disorder (OCD), ageing related metabolic disorders and stress-induced dysfunctions, mania, depression, acute & chronic cases of dementia, etc.) - where chemical & synthetic drugs are not fully effective, different phytomedicinal compounds (phytochemicals from herbal sources) may be successfully utilized with minimum or without side effects even for long term therapy. For the management of different mental & neuronal disorders there is tremendous scope of discovery of specific phytocompounds from medicinal plants. The binding affinity of specific phytochemicals with the gene-products (i.e., specific proteins) of the above disorders using bioinformatic softwares can prove effective for future drug discovery using these phytochemicals [1-3].
Alzheimer’s disease (AD) is a type of dementia that causes problems with memory, thinking and behavior; symptoms usually develop slowly and get worse over time, becoming severe enough that it interferes with daily tasks. The greatest known and studied risk factor is increasing age, and the majority of people with AD are 65 and elder. AD is a progressive disease, where dementia symptoms gradually worsen over a number of years and in its early stages, memory loss is mild, but with late-stage AD, individuals lose the ability to carry on a conversation and respond to their environment. Now, there is a worldwide effort under way to find better ways to treat and fight the disease and to delay its onset. Dementia is the loss of cognitive functioning— thinking, remembering, including reasoning— and behavioral abilities to such an extent that it interferes with a person’s daily life and activities and it ranges in severity from the mildest stage, when it is just beginning to affect a person’s functioning, to the most severe stage, when the person must depend completely on others for basic activities of daily living [4-6].
AD is named after Dr. Alois Alzheimer. In 1906, Dr. Alzheimer noticed changes in the brain tissue of a woman who had died of an unusual mental illness; her symptoms included memory loss, language problems, and unpredictable behavior. After she died, he examined her brain and found many abnormal clumps (also called amyloid plaques) and tangled bundles of fibers (now called neurofibrillary, or tau, tangles); these plaques and tangles in the brain are still considered some of the main features of AD. Another feature is the loss of connections between nerve cells, the neurons in the brain. Neurons transmit messages between different parts of the brain, and from the brain to muscles and organs in the body. Abnormal deposits of proteins form amyloid plaques and tau tangles are found throughout the brain. Once-healthy neurons stop functioning; they lose connections with other neurons and ultimately die. The damage initially appears to take place in the hippocampus, the part of the brain essential in forming memories, and as more neurons die, additional parts of the brain are affected, and they begin to shrink. By the time the final stage of AD has widespread, and brain tissue has shrunk significantly [6-8].
Materials & Methods
For the present study the following medicinal plants have been selected to study the scope & activity of different phyto-compounds using bioinformatic parameter, these are Convolvulus pluricaulis, Morus alba, Bacopa monnieri, Phyllanthus emblica, Gymnema sylvestre, Eclipta alba, Glycyrrhiza glabra, Gymnema sylvestre, Vitex negundo, Picrorhiza kurrooa, Azadirachta indica, Ruta graveolens, Trigonella foenum-graecum, Momordica charantia, Vitex negundo, Terminalia arjuna, Centella asiatica, Bacopa monnieri, Acacia nilotica, Terminalia chebula, Coffea arabica, Sutherlandia frutescens/Bougainvillea spectabilis, Phyllanthus emblica.
Mutated mammalian AMPA implicated as factor causing AD were retrieved from the National Centre for Biotechnology Information (NCBI); templates as retrieved from BLAST were downloaded from PDB. The 3D structure of AMPA was determined by homology modelling. The 3D structures of phytocompounds (from Ayurvedic herbs) were retrieved from various databases. The pharmacophore hypothesis was generated for the existing ligands and the phytocompounds were screened against the generated pharmocophoric hypothesis. Ligands were shortlisted based on their fitness score. Mutations in the AMPA (α-amino-3-hydroxy- 5-methyl-4-isoxazolepropionic acid) are noted as causal factors for many CNS disorders are used in this work [7-9].
▪ AMPA receptor
The α-amino-3-hydroxy-5-methyl-4- isoxazolepropionic acid receptor (also known as AMPA receptor, AMPAR or quisqualate receptor) is a non-NMDA-type ionotropic transmembrane receptor for glutamate that mediates fast synaptic transmission in the central nervous system (CNS) [9-11]. Its name is derived from its ability to be activated by the artificial glutamate analog AMPA. The receptor was first named the “quisqualate receptor” by Watkins and colleagues after a naturally occurring agonist quisqualate and was only later given the label “AMPA receptor” after the selective agonist developed by Honore et al. [10] at the Royal Danish School of Pharmacy in Copenhagen. AMPARs are found in many parts of the brain and are the most commonly found receptor in the nervous system. The AMPA receptor GluA2 (GluR2) tetramer was the first glutamate receptor ion channel to be crystallized. The idea is that AMPARs are trafficked from the dendrite into the synapse and incorporated through some series of signaling cascades. AMPA receptors are responsible for the bulk of fast excitatory synaptic transmission throughout the CNS and their modulation is the ultimate mechanism that underlies much of the plasticity of excitatory transmission that is expressed in the brain [9-12].
In this work the 3D structure of the mutated AMPA receptor responsible for AD and related CNS disorders is modeled & virtually screened against the phytocompounds given.
▪ Methodology
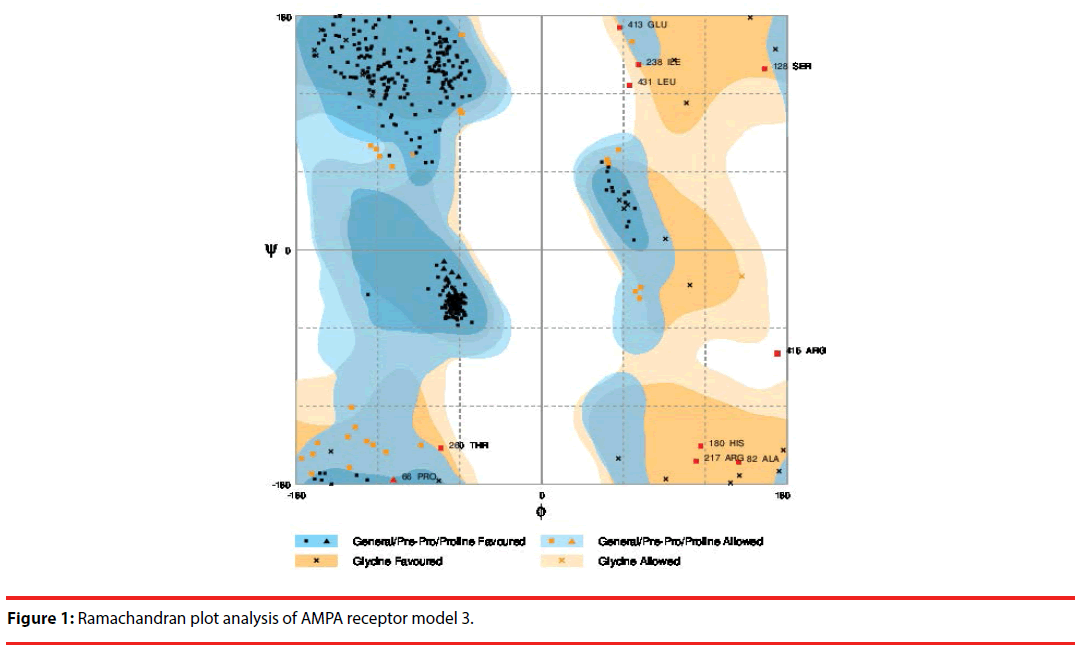
The 3D structure of the AMPA receptor is modelled using modeler software [13]. The AMPA receptor amino acid sequence is downloaded from NCBI; its homologous templates were selected by BLAST. The receptor and their corresponding templates were submitted to modeler software to model their 3D structure. Using Rampage ramachandran plot server the models generated by modeler are analyzed and the best model is selected.
▪ Model quality assessment
Sali and Blundell [13] generated five models. Using Rampage Ramachandran Plot Server, (this stereochemical check was applied to verify if the φ and ψ dihedral angles were in available regions of the Ramachandran plot) the best protein model was selected [14].
Phyto-compounds from traditional ayurvedic herbs
▪ Ligand preparation
The 3d structures of the above phyto-compounds were downloaded from PubChem, a database of chemical molecules maintained by the NCBI and various other online databases.
▪ Generating phase database
Now using Application→Phase→Generate Phase Database module of Maestro software phase database of the phyto-compounds was done [2,15-17].
▪ Selection of ligands for AMPA receptor
Structure-based pharmacophore model is a novel procedure for generating energy-optimized pharmacophore (e-pharmacophores) is based on mapping of the energetic items from the Glide XP scoring function onto atom centers. This was selected by mining the regular features of the three-dimensional structure of AMPA receptor interacting with the known. Phores were selected in the 3D structure of the AMPA receptor at the interaction sites with the known ligands. AMPA receptor with the structural phore information was loaded in the Maestro workspace. Beginning with a ligand-receptor complex structure, we improve the ligand pose, compute the Glide XP scoring items, and map the energies onto atoms. Then, pharmacophore sites were produced, and the Glide XP energies from the atoms that encompass each pharmacophore site were summed. The sites were then graded based on these energies, and the most positive sites were selected for the pharmacophore hypothesis. Finally, these e-pharmacophores were used as queries for virtual screening [2,15-17].
▪ Docking
Docking was performed by Docking server by selecting the best model (model 3) with the ligand selected by pharmacophore modeling, colchicine, to get the docked structure [18].
▪ ADME screening
ADME is an abbreviation in pharmacology and pharmacokinetics for absorption, distribution, metabolism, and excretion. Using Molinspiration server the ADME properties of the selected ligands was determined [19-21]. Molinspiration offers calculation of various molecular properties needed in QSAR and drug design. Molinspiration predicts physically significant descriptors and pharmaceutically relevant properties of molecules. Molinspiration supports cheminformatics for calculation of important molecular properties (logP, polar surface area, number of hydrogen bond donors and acceptors and others), as well as prediction of bioactivity score for the most important drug targets [19,20,21].
Results & Discussion
▪ Homology modelling and model verification
The amino acid sequences of AMPA receptor was downloaded from NCBI (Table 1). Their homologous templates were selected by BLAST (Table 1).
| Receptor | Accession Number | Homologous templates |
|---|---|---|
| AMPA | AAI50210.1 | 5IDEB 4UQQA 4UQ6A |
Table 1: AMPA receptor with its GenBank accession number and homologous templates.
The amino acid sequences of the receptors along with their homologous templates were submitted to modeller software for the generation of the 3D structures of the receptors using the principles of homology modeling [17]. Modeller generated five models for each receptor. The 3D models generated by modeler of AMPA (Table 2) are submitted to Rampage Ramachandran Plot server for model verification [18]. The best 3d AMPA (Figures 1 and 2) model is selected.
| Number of residues in favoured region | Number of residues in allowed region | Number of residues in outlier region | ||
|---|---|---|---|---|
| Model 1 | 391 (90.7%) | 32 (7.4%) | 8 (1.9%) | |
| Model 2 | 393 (91.2%) | 23 (5.3%) | 15 (3.5%) | |
| Model 3 | 393 (91.2%) | 28 (6.5%) | 10 (2.3%) | Selected |
| Model 4 | 395 (91.6%) | 23 (5.3%) | 13(3.0%) | |
| Model 5 | 382 (88.6%) | 37 (8.6%) | 12 (2.8%) |
Table 2: Ramachandran plot analysis of AMPA receptor’s modeler generated models.
▪ Structure-based pharmacophore
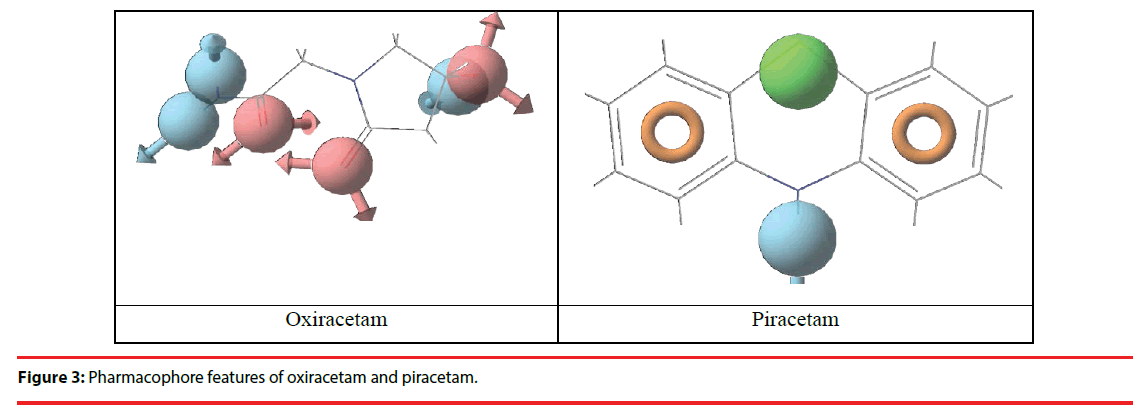
Pharmacophore sites were created in the AMPA receptor (model 3) using the known ligands viz., Oxiracetam [22,23] and Piracetam [24]. The above ligands are established ligands for AMPA receptor. Based on the pharmacophore site information (Figure 3) in the receptor, the unknown ligands in Table 3 were screened.
| Sl. No. | Phyto-Compound under study | Plant Name | Fitness score |
|---|---|---|---|
| Oxiracetam | |||
| 1 | 20-Oxodotriacontanol.1 | Convolvulus pluricaulis | 1.204752 |
| 2 | 1-Deoxynojirimycin.1 | Morus alba | 1.075382 |
| 3 | Bacopaside II.1 | Bacopa monnieri | 1.040053 |
| 4 | beta-Glucogallin.1 | Phyllanthus emblica | 1.021548 |
| 5 | Deacylgymnemic Acid.1 | Gymnema sylvestre | 1.033705 |
| 6 | Eclalbasaponin I.1 | Eclipta alba | 1.137095 |
| 7 | Glycyrrihizin ammonical hydrate.1 | Glycyrrhiza glabra | 1.182755 |
| 8 | Gymnemagenin.1 | Gymnema sylvestre | 0.802587 |
| 9 | Negundoside.1 | Vitex negundo | 0.963057 |
| 10 | Picroside I.1 | Picrorhiza kurrooa | 1.351833 |
| 11 | Picroside II.1 | Picrorhiza kurrooa | 1.186592 |
| 12 | Quercetin dihydrate.1 | Azadirachta indica | 1.004929 |
| 13 | Rutin.1 | Ruta graveolens | 1.178817 |
| 14 | Trigoneoside IVA.1 | Trigonella foenum-graecum | 1.0429 |
| Piracetam | |||
| 15 | vicine.1 | Momordica charantia | 1.351266 |
| 16 | Agnuside.1 | Vitex negundo | 1.315252 |
| 17 | arjunetin.1 | Terminalia arjuna | 1.193699 |
| 18 | arjungenin.1 | Terminalia arjuna | 0.979138 |
| 19 | Asiatic acid.1 | Centella asiatica | 0.882453 |
| 20 | Bacopaside A.1 | Bacopa monnieri | 1.36562 |
| 21 | Catechin 5-O-gallate.1 | Acacia nilotica | 1.099072 |
| 22 | chebulagic acid.1 | Terminalia chebula | 1.077451 |
| 23 | chebulinic acid.1 | Terminalia chebula | 1.048981 |
| 24 | Chlorogenic Acid.1 | Coffea Arabica | 1.25777 |
| 25 | D-Pinitol.1 | Sutherlandia frutescens/ Bougainvilleaspectabilis |
1.599359 |
| 26 | Epicatechin-3-gallate.1 | Tea | 1.098369 |
| 27 | Epigallocatechin 3-gallate.1 | Tea | 1.110839 |
| 28 | Gallic Acid.1 | Phyllanthus emblica | 1.235741 |
(Source: Natural Remedies, Bangalore, India & Satsang Bhesaj Udyan, Deoghar, India)
Table 3: Pharmacophore analysis of phytocompounds.
As per Structure-Based Pharmacophore results phytocompound in Table 3 were selected as the best fitted ligands (Table 3) and the further docking studies (Figure 3) were done using these phyto-compounds.
▪ Molecular docking
AMPA receptor (model 3) was docked with the phytocompound using Docking server. It was seen that AMPA receptor docks with the phytocompounds in Table 4 (Figure 4).
| Ligand Name | Binding Energy | No. of Interactions | Dock | |
|---|---|---|---|---|
| 20-Oxodotriacontanol | -5.00 | 20 | Yes | |
| 1-Deoxynojirimycin | -5.52 | 24 | Yes | |
| Bacopaside II | -8.32 | 20 | Yes | SELECTED |
| beta-Glucogallin | -3.18 | 20 | Yes | |
| Deacylgymnemic Acid | -7.56 | 23 | Yes | |
| Eclalbasaponin I | -8.24 | 25 | Yes | SELECTED |
| Glycyrrihizin ammonical hydrate | -5.51 | 16 | Yes | |
| Gymnemagenin | -6.90 | 15 | Yes | |
| Negundoside | -5.93 | 20 | Yes | |
| Picroside I | -5.93 | 20 | Yes | |
| Picroside II | -6.51 | 30 | Yes | |
| Quercetin dihydrate | -6.10 | 27 | Yes | SELECTED |
| Rutin | -4.79 | 21 | Yes | |
| Trigoneoside IVA | -5.30 | 23 | Yes | |
| Vicine | -4.69 | 18 | Yes | |
| Agnuside | -5.56 | 21 | Yes | |
| arjunetin | -8.26 | 22 | Yes | SELECTED |
| arjungenin | -6.59 | 23 | Yes | |
| Asiatic acid | -7.95 | 23 | Yes | SELECTED |
| Bacopaside A | -5.88 | 15 | Yes | |
| Catechin 5-O-gallate | -5.39 | 24 | Yes | |
| chebulagic acid | -5.86 | 24 | Yes | |
| Gallic Acid | -4.01 | 15 | Yes | |
| chebulinic acid | -6.69 | 19 | Yes | |
| Epigallocatechin 3-gallate | -5.14 | 24 | Yes | |
| Chlorogenic Acid | -4.34 | 9 | Yes | |
| Epicatechin-3-gallate | -6.36 | 36 | Yes | SELECTED |
| D-Pinitol | -4.26 | 8 | Yes |
Table 4: Docking results.
▪ ADME screening
Molinspiration generated the following output (Table 5) for the phytocompounds selected in Table 4.
| miLogP | TPSA | natoms | MW | nON | nOHNH | nrotb | volume | nviolations | |
|---|---|---|---|---|---|---|---|---|---|
| Bacopaside II | 2.36 | 276.15 | 65 | 929.11 | 18 | 10 | 10 | 847.65 | 3 |
| Eclalbasaponin I | 2.40 | 236.06 | 56 | 796.99 | 14 | 9 | 7 | 743.43 | 3 |
| Quercetin dihydrate | 1.68 | 131.35 | 22 | 302.24 | 7 | 5 | 1 | 240.08 | 0 |
| arjunetin | 2.93 | 177.13 | 46 | 650.85 | 10 | 7 | 5 | 619.56 | 2 |
| Asiatic acid | 4.70 | 97.98 | 35 | 488.71 | 5 | 4 | 2 | 487.79 | 0 |
| Epicatechin-3-gallate | 2.54 | 177.13 | 32 | 442.38 | 10 | 7 | 4 | 359.55 | 1 |
Table 5: ADME studies.
Phytocompounds quercetin dihydrate and Asiatic acid were selected as per ADME analysis.
Conclusion
As per Rampage Ramachandran Plot analysis, Model 3 of AMPA receptor is selected as the best model. Further, virtual screening followed by ADME studies it is seen that phytocompounds Quercetin dihydrate from Azadirachta indica and Asiatic acid from Centella asiatica can be successfully used as ligands for AMPA receptor. Further in-vitro receptor binding studies are being performed on the above selected receptor with the selected phytocompounds to establish the efficacy of Quercetin dihydrate and Asiatic acid in treating Alzheimer ’s disease.
Acknowledgement
This work forms a part of SERB-NPDF for Preenon Bagchi, author reference number PDF/2015/000047.
References
- Bagchi P, Anuradha M, Kar A. Ayur-informatics: Establishing an ayurvedic medication for Parkinson's disorder. IJACEBS 4(1), 21-25 (2017).
- Bagchi P, Anuradha M, Kar A. Pharmacophore modeling & docking studies of SNCA receptor with some active phytocompounds from selected ayurvedic medicinal plants known for their CNS activity. Under press with Springer AISC series.
- Bagchi P, Somashekhar R, Kar A. Scope of some Indian medicinal plants in the management of a few neuro-degenetative disorders in silico: A review. Int. J. Public. Ment. Health. Neurosci 2(1), 41-57 (2015).
- Gore M, Bagchi P, Desai NS. Ayur-informatics: Establishing an in silico ayurvedic medication for Alzheimer’s disease. Int. J. Bioinformatics. Res 2(1), 33-37 (2010).
- Burns A, Iliffe S. Alzheimer's disease. BMJ 338(1), b158 (2009).
- Mendez MF. Early-onset Alzheimer's disease: Non-amnestic subtypes and type 2 AD. Arch. Med. Res 43 (8), 677-685 (2012).
- Arnáiz E, Almkvist O. Neuropsychological features of mild cognitive impairment and preclinical Alzheimer's disease. Acta. Neurologica. Scandinavica 179(1), 34-41 (2003).
- Gatz M, Reynolds CA, Fratiglioni L, et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63(2), 168-174 (2006).
- Chang PK, Verbich D, McKinney RA. AMPA receptors as drug targets in neurological disease -Advantages, caveats and future outlook. Eur. J. Neurosci 35(12), 1908-1916 (2012).
- Honore T, Lauridsen J, Krogsgaard-Larsen P. The binding of [3H] AMPA, a structural analogue of glutamic acid, to rat brain membranes. J. Neurochem 38 (1), 173-178 (2012).
- Shi SH, Hayashi Y, Petralia RS, et al. Rapid spine delivery and redistribution of AMPA receptors after synaptic NMDA receptor activation. Science 284 (5421), 1811-1816 (1999).
- Mayer ML. Glutamate receptor ion channels. Curr. Opin. Neurobiol 15 (3), 282-288 (2005).
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol 234(1), 779-815 (1993).
- Laskoswki RA, MacArthur MW, Moss DS. Procheck: A program to check the stereochemical quality of protein structures. J. Appl. Cryst 26(1), 283-291 (1993).
- Schrödinger Suite Protein Preparation Guide, SiteMap 2.4; Glide version 5.6, LigPrep 2.4, QikProp 3.3, Schrödinger, LLC, New York, NY (2010).
- Taha MO, Dahabiyeh LA, Bustanji Y, et al. Combining ligand-based pharmacophore modeling, quantitative structure-activity relationship analysis and in silico screening for the discovery of new potent hormone sensitive lipase inhibitors. J. Med. Chem 51(1), 6478–6494 (2008).
- Singh KHD, Kirubakaran P, Nagarajan S, et al. Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J. Mol. Model 18(1), 39-51 (2012).
- Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminformatics 152009.
- Ertl P, Rohde B, Selzer P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem 43(1), 3714-3717 (2000).
- Lipinski CA, Lombardo F, Dominy BW. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev 23(1), 4-25 (1997).
- Veber DF, Johnson SR, Cheng HY, et al. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem 45(1), 2615-2623 (2002).
- Xiao P, Staubli U, Kessler M. Selective effects of aniracetam across receptor types and forms of synaptic facilitation in hippocampus. Hippocampus 1(4), 373-80 (1991).
- Copani A, Genazzani AA, Aleppo G, et al. Nootropic drugs positively modulate alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-sensitive glutamate receptors in neuronal cultures. J. Neurochem 58(4), 1199-1204 (1992).
- Ahmed AH, Oswald RE. Piracetam defines a new binding site for allosteric modulators of α-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptors. J. Med. Chem 53(5), 2197–2203 (2010).