Research Article - (2018) Volume 8, Issue 6
Circulating Micrornas is a Potential Prognostic Biomarker in Primary Central Nervous System Lymphoma
- *Corresponding Author:
- Xiaoping Xu, Bobin Chen
D"epartment of Hematology, Huashan Hospital, Fudan University, Shanghai, 200040, China
Abstract
Backgrounds and Purposes:
Primary central nervous system lymphoma (PCNSL) is a rare subtype of non-Hodgkin’s lymphoma. The prognosis of PCNSL is poor and heterogeneous. MicroRNA is associated with prognosis of various cancers. Our study is the first time to compare circulating microRNAs of PCNSL patients with different prognosis using miRNA microarray scanning and find novel microRNAs associated with prognosis of PCNSL by further large-scale validation. Materials and
Methods:
From a retrospective cohort, we collected the clinical data of patients who were diagnosed as PCNSL in Huashan Hospital between January 2007 and June 2016. We collected clinical data and blood samples from residual blood routine test of PCNSLs. First, we compared circulating microRNAs between patients with different overall survival (OS) using miRNA microarray scanning. Second, PCR assay was used for miRNAs quantification in blood sample from 6 patients that had long overall survival and other 6 patients that had short overall survival. Further miRNA validation was made by PCR assay in blood sample from 94 patients to validate prognostic value of miRNAs in PCNSLs. The Kruskal-Wallis and Mann-Whitney U tests were used for quantitative parameters and the χ2 square test was used for non-quantitative parameters. Univariate analysis was performed using log-rank test, and survival distributions were analyzed by the Kaplan-Meier curve and log-rank test. Multivariate analysis was done by Cox regression model.
Results:
A total of 90 differentially expressed miRNAs were identified, including 24 upregulated miRNAs and 66 down-regulated miRNAs using miRNA microarray scanning. Then PCR assay was used for 10 differentially expressed miRNAs quantification in blood sample from 6 patients that had long overall survival and 6 patients that had short overall survival. Mir-21, mir-129-5p, mir-144-5, mir-363-5p, mir-409-3p, mir-1246, mir-1299 and mir-1825 showed no differences between 2 groups, while the expression of mir-455-3P and mir-940 between 2 groups are significantly different (P<0.05). Further validation in 94 patients showed median OS in miR-940 overexpression group was 91 months and in miR-940 low-expression group was 28 months (P=0.0005), while median PFS in miR-940 overexpression group was 25 months and in miR-940 low-expression group was 16 months (P=0.0292). Multivariate analysis of COX model showed miR-940was independent factors for OS and PFS.
Conclusion:
Circulating mir-940 had prognostic value in PCNSL and was an independent prognostic factor for PCNSL.
Keywords
Primary central nervous system lymphoma, microRNA, Prognosis
Introduction
Primary central nervous system lymphoma (PCNSL) is a rare extranodal form of non- Hodgkin lymphoma (NHL), which occurs intracerebrally or intraocularly, with poor longterm survival. These lymphomas exclusively arise in the central nervous system (CNS) such as the brain parenchyma [1], spinal cord, eyes and/ or cranial nerves. Histologically, approximately 90% of PCNSL are diffuse large B cell lymphoma (DLBCL). The prognosis of PCNSL is poor and heterogeneous and PCNSL patients are often lack of specific, sensitive, noninvasive manifestations to diagnosis and prognosis. MiRNAs are small regulatory, noncoding RNAs. It is believed that miRNAs primarily affect the stability of mRNA and/or the initiation and progression of protein translation [2-6]. Even though the biological function of miRNA is yet to be fully understood, it has been shown that miRNAs play multiple roles in cancer genesis, development, invasion and metastasis [7-9]. However, there are few studies on miRNAs involved in prognosis of PCNSL.
In the current study, we established retrospective cohort of PCNSL patients. Patients with poor overall survival (OS) were classified as the case group, whereas patients with good OS were classified as the control group. We studied the different microRNA gene expression of patients in the case and control groups to gain insight into the specific miRNA associated with PCNSL prognosis.
Materials and Methods
▪ Patients and ethics statement
A total of 151 patients newly diagnosed with PCNSL between July 2007 and June 2016 in Huashan Hospital were retrospectively enrolled in this study, with the enrollment criteria of HIV-negative and non-immunosuppressionrelated PCNSL. These patients were pathologically reconfirmed according to the 2008 WHO classification of lymphomas. Clinico pathological data were collected, including clinical presentations, diagnoses, regimens, therapeutic outcomes, second-line treatments, toxicity reactions, and survival durations. Signed informed consent was obtained from all patients or guardians. All patients underwent gadolinium-enhanced cranial magnetic resonance imaging (MRI) scan and/or positron emission tomography/computed tomography (PET/CT) before and after chemotherapy to evaluate the response to treatment. Serum lactate dehydrogenase (LDH), total creatinine clearance (CCr), B ultrasound of lymph nodes and abdomen, slit-lamp examination, as well as bone marrow biopsy and aspiration were also conducted in all patients.
▪ RNA extraction
The total RNA from peripheral blood mononuclear cells was extracted using TRIzol (Invitrogen, Paisley, U.K.) according to the manufacturer’s protocol. Aliquots of the RNA samples were conserved for quality evaluation using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
▪ Array hybridization, data analyses and target prediction of miRNAs
To assess the level and composition of miRNA, miRNA arrays from Agilent Technologies were used. The RNA samples were labeled and processed according to manufacturer’s recommended protocols. In brief, ≈100 ng of total RNA was dephosphorylated with calf intestinal alkaline phosphatase, followed by denaturing with heat in the presence of dimethyl sulfoxide (DMSO). A cyanine dye, cyanine3- cytidine bisphosphate (pCp), was then joined to the dephosphorylated single-stranded RNA (including miRNA) by T4RNA ligase. Micro BioSpin 6 columns (Bio-Rad) were used to remove any unincorporated cyanine dye from the samples. The purified labeled miRNA probes were hybridized to 8 × 15 K human miRNA microarrays in a rotating hybridization oven at 20 rpm for 20 h at 55°C. After hybridization, the arrays were washed in Agilent gene expression (GE) Wash Buffer 1 with TritonX-102, followed by Agilent GE Wash Buffer 2 with Triton X-102. After washing, all slides were immediately scanned at 5 μm resolution by using a PerkinElmer ScanArray Express array scanner. The resulting images were quantified by using Agilent’s Feature Extraction software. The differentially expressed miRNAs were identified using Agilent miRNA gene arrays protocol. To increase the reliability of the data, miRNA species with hybridization intensities<1.5 times the average hybridization intensity (mean) were excluded from analysis. The miRNA clustering analysis was performed with the Hierarcical clustering algorithm provided in Multi Experiment Viewer, MeV4.0 software package (www.tm4.org/mev.html). After screening the differentiated miRNA, we predicted the targeting genes using targets can software. Sites with PCT>0.75 are with higher probability of preferential conservation.
▪ Statistical analysis
Descriptive and explorative data analyses of all parameters were done. Quantitative parameters were tested for normal distribution. The Kruskal- Wallis and Mann- Whitney U tests were used for quantitative parameters; the χ2 square test and the Fisher’s test for non-quantitative parameters. Survival and follow-up were calculated from the date of PCNSL diagnosis to the time of death, recurrence, progression, or last followup. Univariate analysis was performed using log-rank test, and survival distributions were analyzed by the Kaplan-Meier curve and logrank test. Multivariate analysis was done by Cox regression model. All tests were two-sided and P<0.05 was considered to be statistically significant. All statistical analyses were performed with Stata (Version 12.0) and Graph Pad Prism (Version 6.0).
Results
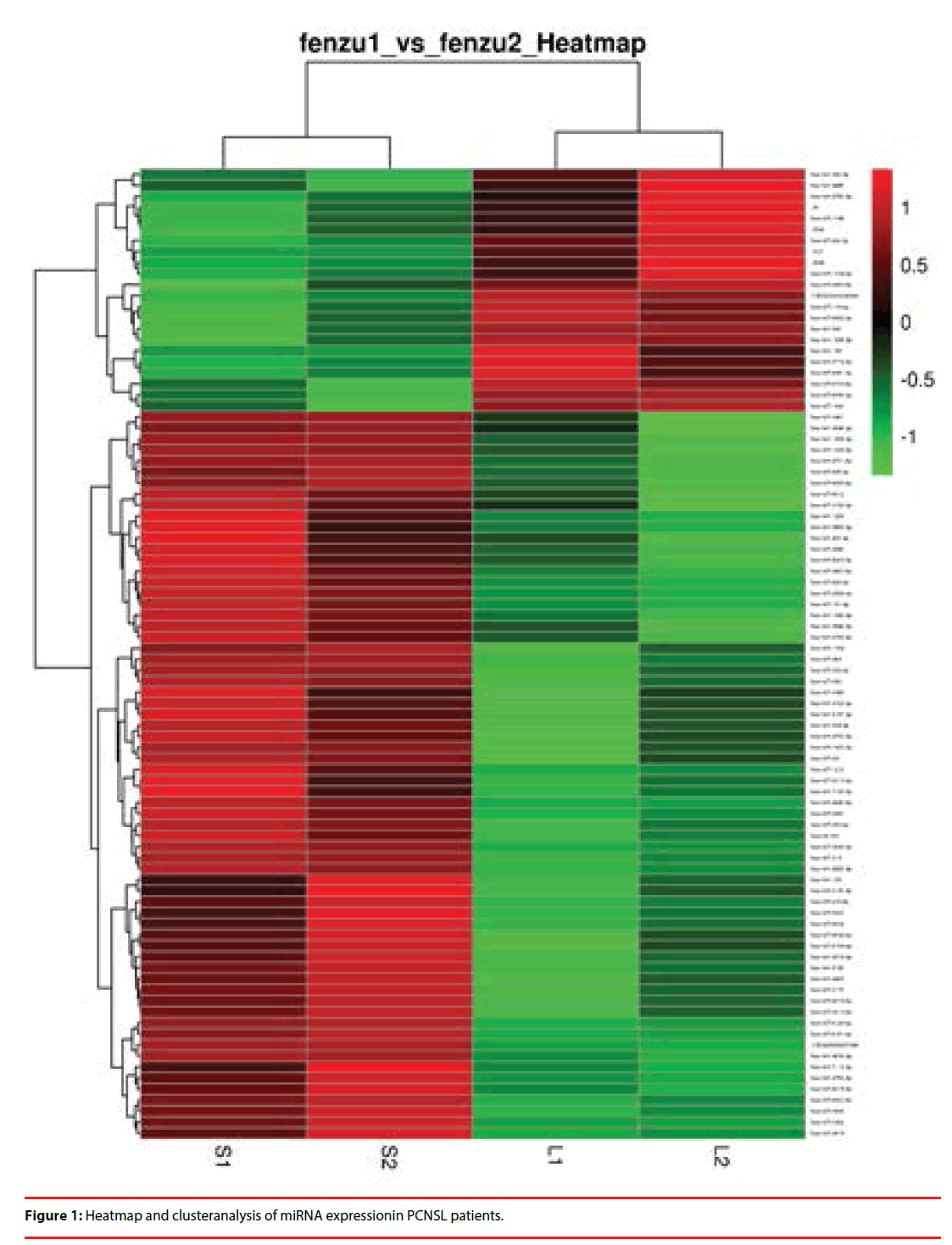
From the above retrospective cohort of PCNSL patients, paired patients were chosen for miRNA microarray screening according to age, gender, ECOG, Pathology, LDH level, CSF protein level, and Involvement of deep structures (Table 1). Long OS included patients with more than 3 years OS, while short OS included patients with less than 1 years OS. We adapted a microarray platform from Agilent to profile the miRNA spectra. Excluding any miRNA with hybridization intensity <1.5 times the global mean intensity, there were 90 miRNAs to be significantly differentially expressed between two group, including 66 down-regulated miRNA and 24 upregulated miRNA (Figure 1). The main characteristics of paired patients for miRNA microarray assay are listed in Table 1.
| Characteristics | Long OS | Short OS |
|---|---|---|
| Age,years(`x±SD) | 54±3.5 | 50±6.4 |
| Sex | ||
| Male | 2 | 2 |
| Female | 1 | 1 |
| ECOG | ||
| 0-1 | 2 | 2 |
| ≥1 | 1 | 1 |
| Pathology | ||
| DLBCL | 3 | 3 |
| Elevated serum LDH level | 0 | 0 |
| Elevated CSF protein level | 1 | 1 |
| Involvement of deep structures | 1 | 1 |
| Abbreviations: ECOG: Eastern Cooperative Group Score;DLBCL,diffuse large B-cell lymphoma;LDH,lactate dehydrogenase; CSF,cerebrospinal fluid. | ||
Table 1: The clinical characteristics of paired patients for miRNA microarray assay.
We selected miRNAs with p values of expressional differences less than 0.05 and/or >2-fold change between two group in miRNA array analysis. The heat map generated by using heat map software shows changes in the expression of these miRNAs in short OS group compared with long OS group. miRNAs whose expression is higher in short OS group are shown in red and those that are lower in green. The color key represents the values that are scaled to have a mean of zero and standard deviation of one. The identities of miRNAs are listed on the right, whereas the sample names are on the bottom.
Cluster analysis of above 90 miRNAs resulted in a subset of 10 differentially expressed miRNAs, includingmir-21, mir-129-5p, mir-144-5, mir- 363-5p, mir-409-3p, mir-455-3p, mir-940, mir- 1246, mir-1299 and mir-1825. Then validation of these 10 differentially expressed miRNAs was made in 6 patients with long OS and 6 patients with short OS using PCR assay. The main characteristics of paired patients for miRNA microarray assay are listed in Table 2. Mir-21, mir-129-5p, mir-144-5, mir-363-5p, mir-409- 3p, mir-1246, mir-1299 and mir-1825 showed no differences between 2 groups, while the expression of mir-455-3P and mir-940 between 2 groups are significantly different (P<0.05) (Table 3).
| Characteristics | Long OS | Short OS | z | p |
|---|---|---|---|---|
| Sex(male/female) | 4/2 | 5/1 | 0.638 | 0.5233 |
| Age(years) | 52±7.22 | 58±4.67 | 1.771 | 0.0766 |
| OS(months)(mean) | 87(73-108) | 8(1~11) | -2.892 | 0.0038 |
Table 2: The clinical characteristics of 12 patients for validation of 10 differentially expressed miRNAs.
| miRNAs | Short OS Median(range) |
Long OS Median(range) |
U | P |
|---|---|---|---|---|
| mir-21 | -6.97(-8.06~-5.87) | -6.12(-7.08~-5.16) | 11 | 0.31 |
| mir-129-5p | -18.03(-20.49~-15.57) | -15.86(-16.82~-14.90) | 7 | 0.09 |
| mir-144-5p | -19.22(-21.46~-16.97) | -17.06(-18.42~-15.70) | 9 | 0.18 |
| mir-363-5p | -19.42(-21.55~-17.29) | -17.45(-21.55~-17.29) | 5 | 0.051 |
| mir-409-3p | 1.72(-0.76~4.21) | 2.95(2.31~3.59) | 15 | 0.7 |
| mir-455-3p | -18.53(-20.59~-16.46) | -16.49(-17.29~-15.69) | 4 | 0.03 |
| mir-940 | -18.74(-20.59~-16.46) | -16.53(-17.29~-15.69) | 5 | 0.04 |
| mir-1246 | -10.24(-12.48~-8.00) | -8.423(-9.13~-7.71) | 7 | 0.09 |
| mir-1299 | -17.24(-19.42~-15.07) | -15.86(-17.70~-14.86) | 11.5 | 0.33 |
| mir-1825 | -18.04(-20.24~-15.83) | -16.14(-17.11~-15.17) | 9 | 0.18 |
Table 3: Expression of 10 differentially expressed circulating miRNAs in 12 patients (6 Short OS patients and 6 long OS patients).
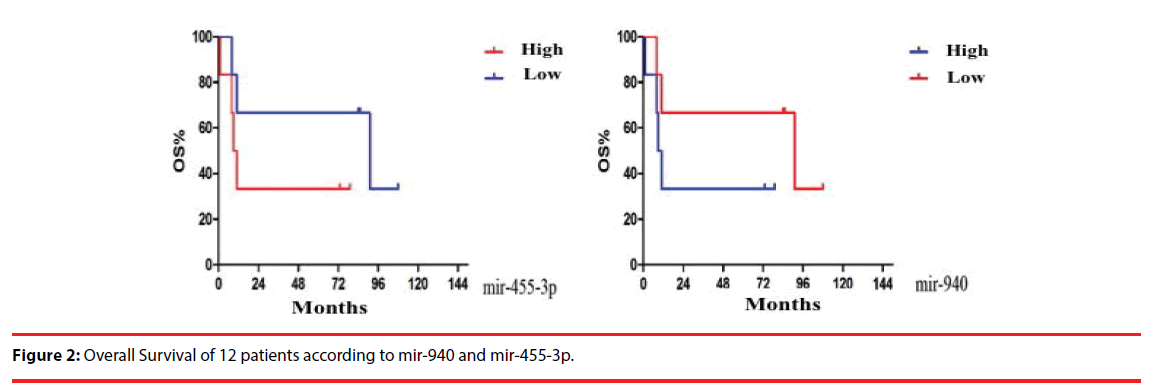
Kaplan-Meier curves of 12 patients showed median OS in miR-455-3p overexpression group and low-expression group was 10 months and 91 months, respectively. Median OS in miR-940 overexpression and low-expression group was 91 months and 10 months, respectively (P ≥ 0.05) (Figure 2).
Further validation of mir-455-3p and mir- 940 was made by Kaplan-Meierin 94 patients. Median OS in miR-455-3p overexpression group was 28 months and in miR-940 low-expression group was 56 months (P=0.12). And median PFS in miR-455-3p overexpression group was 23 months and in miR-940 low-expression group was 16 months (P=0.16) (Table 4).
| Highexpression(months) | Lowexpression(months) | X2 | P | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mir-455-3p | OS | 28 | 56 | 1.913 | 0.17 | |||||||||||||
| PFS | 23 | 18 | 2.00 | 0.16 | ||||||||||||||
Table 4: Overall Survival and PFS of 94 patients according to mir-455-3p expression.
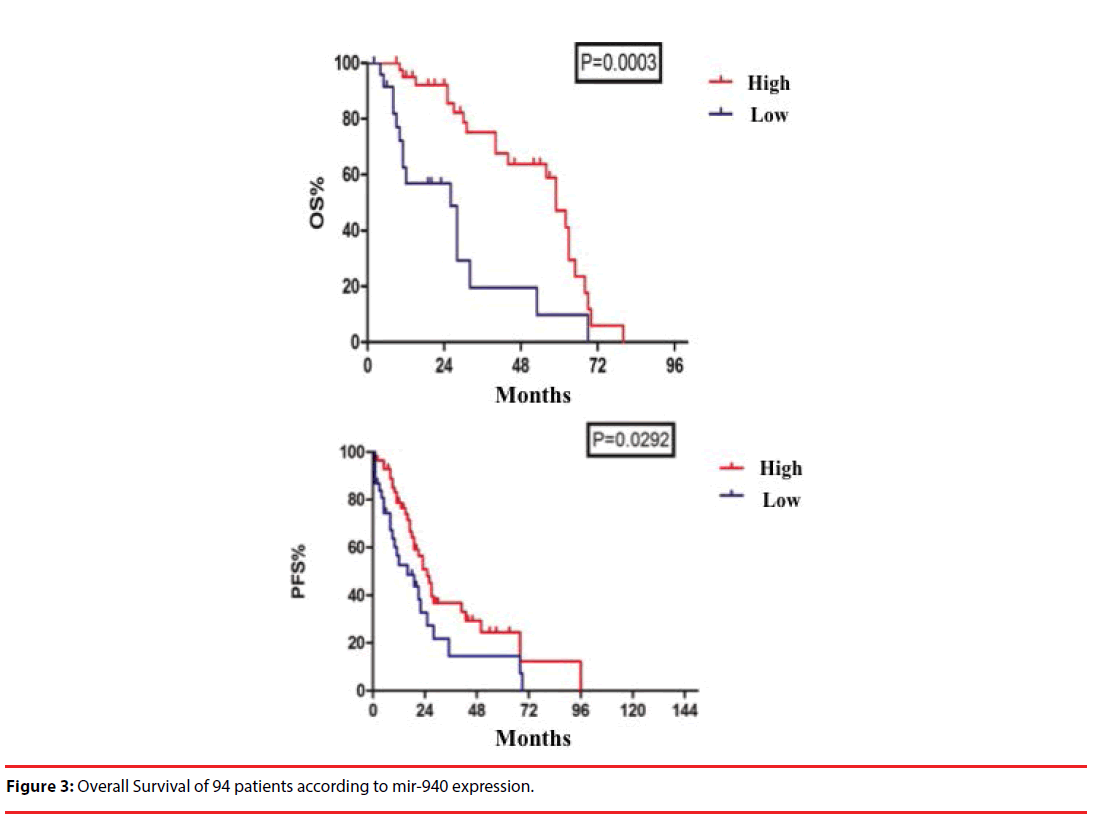
Median OS in miR-940 overexpression group was 91 months and in miR-940 low-expression group was 28 months (P=0.0003). And median PFS in miR-940 overexpression group was 25 months and in miR-940 low-expression group was 16 months (P=0.0292) (Figure 3).
Multivariate analysis included risk prognostic factor of PCNSL in univariate analysis by Logrank, including age, ECOG, KPS, Standard Uptake Value (SUV) of PET-CT, Sr and mir- 940 (Data not shown). Table 5 showed ECOG and mir940 were independent risk prognostic factors of PCNSL patients.
| Factors | B | SE | Wald | Sig. | OR | 95.0% CI |
| age | -0.39 | 0.292 | 1.789 | 0.181 | 0.677 | 0.382-1.199 |
| ECOG | -0.65 | 0.292 | 4.959 | 0.026 | 0.522 | 0.295-0.925 |
| KPS | 0.274 | 0.279 | 0.966 | 0.326 | 1.315 | 0.762-2.27 |
| SUV | -1.842 | 0.997 | 3.411 | 0.065 | 0.159 | 0.022-1.119 |
| Sr | 0.661 | 1.316 | 0.253 | 0.615 | 1.937 | 0.147-25.537 |
| mir940 | -1.995 | 1.007 | 3.925 | 0.048 | 0.136 | 0.019-0.979 |
Table 5: Multivariate analysis of prognostic risk factors of PCNSL patients.
Discussion
The prognosis of PCNSL is poor and heterogeneous. PCNSL patients are often lack of specific, sensitive, noninvasive manifestations to diagnosis and prognosis. Recently, research studied the prognosis value of clinical, biochemical and immune examination and found that some factors are involved, including the patient’s age, performance status (PS), serum LDH levels, cerebrospinal fluid protein concentration, deep brain and etc. MiRNAs are small regulatory, noncoding RNAs and play multiple roles in cancer genesis, development, invasion and metastasis. Compared with mRNA, miRNAs are more stable and specific so as to be an ideal tumor marker. However, there are few studies on miRNAs involved in prognosis of PCNSL.
In the current study, it is the first time to comparing circulating microRNAs of PCNSLs that had different prognosis using miRNA microarray scanning and finding novel microRNAs associated with prognosis of PCNSL by further large-scale validation. We identified 90 miRNAs to be significantly differentially expressed between two groups with long and short OS. We finally screened a subset of 10 miRNAs for further validation in 6 patients with long OS and 6 patients with short OS, showing the expression of mir-455- 3P and mir-940 between 2 groups are significantly different (P<0.05) (Table 3).
Recently study showed mir-455-3p can promote prostate cancer cell invasion and metastasis by targeting the tumor suppressor gene EI24, and high expression of mir-455-3p is associated with poor prognosis [10]. In colon cancer cell lines, mir-455-3p induces apoptosis and inhibits cell proliferation by targeting the tumor suppressor gene HCT116 [11]. In glioblastoma cells resistant to temozolomide, mir-455-3p was significantly higher and overexpression of mir-455-3p showed a cytotoxic effect when temozolomide was used [12]. High expression of mir-455-3p is also observed in basal cell carcinoma of the skin [13]. In particular, in DLBCL patients with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone) treatment, high expression of blood circulating mir-455-3p is associated with high relapse [14]. Our study showed patients with high mir-455-3p expression had short OS and long PFS although there is no statistic significance.
Expression of mir-940 was found lower in prostate cancer tissues than normal tissues, and the mechanism may be that mir-940 can inhibit cancer cell migration and invasion by regulating the migration and invasion of enhanced gene 1 (Migration and invasion enhancer 1, MIEN1) [15]. In pancreatic ductal adenocarcinoma, high expression of mir-940 also inhibited pancreatic ductal adenocarcinoma cell growth, whereas low mir-940 expression was associated with a low survival rate [16]. However, the results of other study in gastric cancer are different from those of prostate cancer and pancreatic ductal adenocarcinoma, that high expression of mir-940 is associated with poor prognosis. The authors suggest that mir-940 can enhance the ability of invasion and metastasis of gastric cancer by down-regulating the expression of zinc finger transcription factor 24 (Zinc, Finger, Transcription, Factor, 24, ZNF24) [17]. In patients with hepatocellular carcinoma, low expression of mir-940 is associated with poor prognosis because he mir-940 can inhibit cells growth and induce apoptosis of hepatocellular carcinoma by inhibiting estrogen receptor gamma (Estrogen-related receptor, gamma, ESRRG) [18]. A study by Xie et al. shows that mir-940 expression is up regulated in saliva of patients with pancreatic cancer and can be used in the diagnosis of early pancreatic cancer [19]. Our study showed patients with low mir-940 expression had long OS although there is no statistical significance (P=0.16).
In all, our study showed circulating mir-940 is a potential prognostic biomarker for PCNSL; nevertheless, our research has intrinsic limitations in consideration that it is a retrospective, singleinstitution analysis. The matching of the two patient cohorts are suboptimal by reason that patients were not prospectively assigned to the different treatment groups using prognostic factors for randomization. Thus, future wellcontrolled prospective studies are required to confirm the current data.
Authorship
Yan MA and Dina Suolitiken analyzed patients’ data and wrote the manuscript. Xiaoping Xu and Bobin Chen reviewed and supervised research and approved the manuscript. Lu Zhang, Hui Kang, Zhiguang Lin and Qian Wang participated in the implementation of the treatment.
Conflict of Interest
The authors declare that they have no conflict of interest.
Acknowledgement
The work was supported by grants from China National Clinical Key Subject Construction Foundation (Oncology Department, 2013), Shanghai Pujiang Program (Grant No.16PJD011), Shanghai Hosptial Development Center (Grant No.16CR2043B) and Shanghai Municipal Commission of Health and Family Planning (Grant No.201640132).
References
- Nayak L, Iwamoto FM, LaCasce A, et al. PD-1 blockade with nivolumab in relapsed/refractory primary central nervous system and testicular lymphoma. Blood 129(1), 3071-3073 (2017).
- Sakaguchi T, Yoshino H, Yonemori M, et al. Regulation of ITGA3 by the dual-stranded microRNA-199 family as a potential prognostic marker in bladder cancer. Br. J. Cancer 116(1), 1077-1087 (2017).
- Li Z, Chen P, Su R, et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood 126(1), 2005-2015 (2015).
- Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U S A105(1), 10513-10518 (2008).
- Emmrich S, Engeland F, El-Khatib M, et al. miR-139-5p controls translation in myeloid leukemia through EIF4G2. Oncogene 35(1),1822-1831 (2016).
- Trevisani F, Ghidini M, Larcher A, et al. MicroRNA 193b-3p as a predictive biomarker of chronic kidney disease in patients undergoing radical nephrectomy for renal cell carcinoma. Br. J. Cancer 115(1),1343-1350 (2016).
- Itkin T, Kumari A, Schneider E, et al. MicroRNA-155 promotes G-CSF-induced mobilization of murine hematopoietic stem and progenitor cells via propagation of CXCL12 signaling. Leukemia 31(1),1247-1250 (2017).
- Danilova OV, Paiva C, Kaur P, et al. MIR21 is differentially expressed in the lymphoid tissue and modulated by stromal signalling in chronic lymphocytic leukaemia. Br. J. Haematol 170(1), 272-275 (2015).
- Sierzega M, Kaczor M, Kolodziejczyk P, et al. Evaluation of serum microRNA biomarkers for gastric cancer based on blood and tissue pools profiling: the importance of miR-21 and miR-331. Br J Cancer 117(1), 266-273 (2017).
- Li Z MQ, Pan A, Wu X, et al. MicroRNA-455-3p promotes invasion and migration in triple negative breast cancer by targeting tumor suppressor EI24. Oncotarget 8(1), 19455-19466 (2017).
- Zheng J, Lin Z, Zhang L, et al. MicroRNA-455-3p Inhibits Tumor Cell Proliferation and Induces Apoptosis in HCT116 Human Colon Cancer Cells. Med. Sci. Monit 22(1), 4431-4437 (2016).
- Ujifuku K, Mitsutake N, Takakura S, et al. miR-195, miR-455-3p and miR-10a( *) are implicated in acquired temozolomide resistance in glioblastoma multiforme cells. Cancer. Lett 296(1), 241-248 (2010).
- Sand M, Skrygan M, Georgas D, Hahn SA, et al. Expression of microRNAs in basal cell carcinoma. Br. J. Dermatol 167(1), 847-855 (2012).
- Song G, Gu L, Li J, et al. Serum microRNA expression profiling predict response to R-CHOP treatment in diffuse large B cell lymphoma patients. Ann. Hematol 93(1), 1735-1743 (2014).
- Rajendiran S, Parwani AV, Hare RJ,et al. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol. Cancer 13(1), 250 (2014).
- Song B, Zhang C, Li G, et al. MiR-940 inhibited pancreatic ductal adenocarcinoma growth by targeting MyD88. Cell. Physiol. Biochem 35(1), 1167-1177 (2015).
- Liu X, Kwong A, Sihoe A, et al. Plasma miR-940 may serve as a novel biomarker for gastric cancer. Tumour. Biol 37(1), 3589-3597 (2016).
- Yuan B, Liang Y, Wang D, et al. MiR-940 inhibits hepatocellular carcinoma growth and correlates with prognosis of hepatocellular carcinoma patients. Cancer. Sci 106(1), 819-824 (2015).
- Xie Z YX, Gong B. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer. Prev. Res (Phila) 8(1), 165-173 (2015).