Research Article - Neuropsychiatry (2018) Volume 8, Issue 1
Association of Small Ubiquitin-related Modifier 3 Gene Polymorphisms with Alzheimers disease in Korean Population
- Corresponding Author:
- Won-Cheoul Jang
Department of Chemistry, College of Natural Science, Dankook University, Cheonan 31116, South Korea
Tel: +82-41-529-6256
Fax: +82-41-559-7860
Abstract
Abstract
Background: Sumoylation is an important post-translation modification that regulates various cellular pathways where small ubiquitin-related modifiers (SUMOs) are covalently linked to lysine residues of cellular target proteins. SUMO has proven to be linked to neuronal development and associated
with pathogenesis of Alzheimer’s disease (AD). Among the SUMO family, SUMO1 and 2 gene polymorphisms were significantly associated with risk of AD. However, genetic study of SUMO3 gene in AD has not been reported yet.
Methods: The association between SUMO3 gene polymorphisms and risk of AD was investigated in Korean population consisted of 144 AD patients and 329 healthy controls. The five tag SNPs within the SUMO3 gene were selected based on observed linkage disequilibrium (LD) and were used for
genotyping. In this case-control study, statistical analysis using logistic regression evaluated to odds ratio (OR) and 95% confidence interval (95% CI) for the relationship between these polymorphisms and risk of AD.
Results: The CT genotype of rs235337 was significantly associated with a decreased risk of AD (OR: 0.455, 95% CI: 0.211-0.979, p-value: 0.044) in female group. In contrast, the rs2838694 GG genotype was related with a highly increased risk of AD (OR: 26.88, 95% CI: 1.197-603.5, p-value: 0.038) in female group. After conducting subgroup analysis of the selected five tag SNPs using Apolipoprotein E (ApoE) ε4 status, the CT genotype of rs235337 with a decreased risk of AD (OR: 0.355, 95% CI: 0.153- 0.827, p-value: 0.016), and the GG genotype of rs2838694 with a highly increased risk of AD (OR:
36.71, 95% CI: 1.579-853.3, p-value: 0.025) were investigated in the ApoE ε4 non-carrier group. In ApoE ε4 carrier status, all tag SNPs were not associated with a risk of AD.
Conclusion: This is a case-control genetic association study to investigate whether the risk of AD is linked to SUMO3 polymorphisms in a Korean population. The two tag SNPs, rs235337 and rs2838694, within the SUMO3 gene showed that they have potential genetic risk factors for AD in a Korean
population. Furthermore, investigation of genetic association study will be useful to determine the true susceptibility of SUMO3 polymorphism to AD.
Keywords
Alzheimer`s disease, Small ubiquitin-related modifier 3, Polymorphism
Introduction
Alzheimer’s disease (AD) is most common types of dementia causing problems including memory loss, solving problems, confusion of time or place, and poor judgement [1]. AD has become a major public health crisis throughout the world since approximately 35.6 million people were estimated to have dementia in 2010, and that number is projected to increase to 65.7 million by 2030 and 115.4 million by 2050 [2]. AD is characterized by senile plaques and neurofibrillary tangles (NFT) in hippocampus and cerebral cortex of brain [3,4]. Amyloid β (Aε), major element of senile plaques, is generated by cleavage of amyloid β precursor protein (APP) and its aggregation caused by pathogenesis of AD [4]. In addition, hyperphosphorylated form of tau protein, major element of NFT, is expressed in the neocortex, hippocampus and amygdala of AD brain [5].
Sumoylation, the covalent attachment with small ubiquitin-like modifier (SUMO) protein family and lysine residues in target protein, is reversible post-translation modification involved in various cellular processes [6]. In human, SUMO proteins indicate the four distinct types including SUMO1, 2, 3 and 4. These SUMO proteins, are similar to three dimensional structure of ubiquitin protein, expressed ubiquitously, but it is not clear SUMO4 is expressed in cells [7,8]. It is known that sumoylation cycle for covalent modification to target protein is processed by the three distinct steps. SUMO protein which is cleaved by sentrin-specific protease (SENP) binds to SUMO-specific activating enzyme heterodimer (SAE1 and SAE2) using ATP, and subsequently, SUMO protein is transferred to the E2-conjugating enzyme UBC9 and conjugated to the lysine of substrate that is defined by the consensus motif ΨKXE/D. Finally, isopeptide bond is formed between SUMO and target protein by the E3 ligase [6]. Previous studies have provided that sumoylation was related with several human disease pathogenesis. For example, overexpression of the UBC9 gene encoding the SUMO-conjugating enzyme was associated with cancer cell growth [9], and the overexpression of SUMO specific protease 1 (SENP1) in prostate promotes neoplasia development [10]. Moreover, a number of studies has provided that sumoylation of proteins has been implicated in the neurodegenerative disease associated with huntingtin (Huntington’s disease), Tau (Parkinson’s and Alzheimer’s disease), α-synuclein (Parkinson’s disease), SOD1 (Amyotrophic lateral sclerosis) and APP (Alzheimer’s disease) [11].
Based on the evidence available to date, there are some scientific studies of SUMO3 supporting that there is a potential relationship between APP processing and SUMO3 because APP is known to be associated with pathogenesis of AD. It has reported that increasing SUMO3 sumoylation is correlated with the negative regulation of Aβ production in neuronal cells and vice versa, which illustrates a regulatory mechanism in APP amyloidogenesis by sumoylation [12]. In addition, SUMO3 protein has been reported to be significantly associated with AD pathogenesis because increased levels of SUMO3 significantly produces both Aβ40 and Aβ42 peptides in HEK-293 cells compared with transfected controls [13]. SUMO3 sumoylation requires the C-terminal di-glycine residues (G92A, G93A), and these residue mutants can cause higher level of N-terminal ADAM-cleaved AβPP fragment (α-NTF) and inversely lower levels of N-terminal BACE1-cleaved AβPP fragment (β-NTF) which generated Aβ in APP processing [12]. Accumulating evidence has elucidated that SUMO proteins and SUMO related proteins play a critical role in AD pathogenesis; therefore, this case-control study was designed to help determine if SUMO3 polymorphisms were associated with a risk of AD in a Korean population. Here, we investigated the associations of the five tag SNPs based on linkage disequilibrium (LD) analysis with AD risk in a Korean population consisted of 144 AD patients and 329 healthy controls and examined further stratified analyses by Apolipoprotein E (ApoE) ε4 status. This is the first population-based case-control study focusing on the association between SUMO3 and AD risk [14].
Materials and Methods
▪ Subjects
The study subjects consisted of 144 AD patients and 329 healthy controls without dementia. The mean age of the AD group (119 females, 25 males) was 79.82 ± 7.02 years and that of the controls (164 females, 165 males) 68.85 ± 6.09 years. The average educational level of the AD group (2.92 ± 3.59 years) was lower than that of the control group (8.29 ± 5.29 years). We define education level by years of education. We recruited AD patients from a dementia clinic of Dankook University Hospital. The cognitively normal control participants were community-dwelling elders who had participated in the Korean Longitudinal Study on Cognitive Aging and Dementia (KLOSCAD) and the Nationwide Dementia Screening and Registration Program (NDSRP) [15]. All subjects underwent standardized clinical interviews and neurological and physical examinations, administered by geriatric psychiatrists with advanced training in neuropsychiatry and dementia research. We used the Korean version of the protocol of the clinical assessment battery established by the Consortium to Establish a Registry for Alzheimer’s disease (CERAD-K) to diagnose dementia [16]. Dementia was diagnosed using the criteria of the Diagnostic and Statistical Manual of Mental Disorders, Fourth Edition (DSM-IV), and probable or possible AD was diagnosed using the criteria of the National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s disease and Related Disorders Association (NINCDS-ADRDA) [17]. All participants were fully informed about the aims of the study and each participant, or his/her legal guardian, gave written informed consent [18].
▪ Assessment of neuropsychological functions
Global cognitive function was evaluated via Mini-Mental Status Examination (MMSE) [16,19]. To assess memory, the word list memory test (WLMT), the word list recall test (WLRT), and the word list recognition test (WLRcT) of the Korean version of the CERAD Neuropsychological Assessment Battery (CERAD-K-N) were administered. Language function was evaluated using the verbal fluency test (VFT). The 15-item modified Boston Naming Test (mBNT) from the CERAD-K-N was also employed [16,19]. Visuospatial function was evaluated using the constructional praxis test (CPT) of the CERAD-K-N. Frontal function was evaluated using the trail making test A and B of the CERAD-K-N [16,18,19].
▪ Genotyping analysis
Five tag single nucleotide polymorphisms (SNPs) retrieved from LD information were retrieved for genotyping using 144 AD patients and 329 healthy controls using PCR and Sanger sequencing. The PCR primers were manually designed (Table 1), and all PCR reactions of the five tag SNPs were performed on a C1000 Thermal Cycler (Bio-Rad, Hercules, CA, USA). Total volume of 25 μL PCR products was consisted of 2.5 mM MgCl2, 2.5 mM of each dNTP, 100 μM of each primer, 50 ng template DNA, and 1 unit of Taq DNA polymerase. The PCR protocol was as follows: initial denaturation at 94°C for 5 min; 33 cycles of 94°C for 30 s, 61°C for 30 s, and 72°C for 45 s; and a final extension at 72°C for 3 min. Genotyping of PCR products was directly carried out on BigDye® Terminator v3.1 cycle sequencing kit on an ABI 3730XL DNA sequencer (Applied Biosystems, Foster City, CA, USA).
| Sequence (5'->3') | GC (%) | Tm (°C) | Product size (bp) | ||
|---|---|---|---|---|---|
| rs235337 (C>T) | Forward | CGTCTTTGCGTGCTCCTCTC | 60 | 58.4 | 315 |
| Reverse | GTGGGAGTAGGTGCACACTG | 60 | 57.8 | ||
| rs3827259 (A>G) | Forward | CGCTTCCTCACCTGGGTTTC | 60 | 58.1 | 290 |
| Reverse | TGCCTGACACCACTGCTTTC | 55 | 58.4 | ||
| rs2838694 (C>G) | Forward | GTCCATCAGACCACCAGTGT | 55 | 56.8 | 258 |
| Reverse | TTCTGCGTCCTGAGGGAATG | 55 | 57.1 | ||
| rs2838697 (G>T) | Forward | CATCAACCTGAAGGTGGCCG | 60 | 58.7 | 296 |
| rs13050872 (C>G) | Reverse | AAAAGGCCATGGGGGACAAG | 55 | 58.2 |
Table 1: Primer set for five tagSNPs used in this study.
▪ Statistical analysis
Tag SNPs were identified based on pairwise LD with the aid of National Institute of Environmental Health Sciences web software (https://snpinfo.niehs.nih.gov) with HapMap database. All statistical analyses were performed using SPSS version 23.0 software (SPSS Inc., Chicago, IL, USA). The chi-squared test was used to explore whether genotype distributions in the controls were in Hardy-Weinberg Equilibrium (HWE). The association between tag SNPs and AD risk was investigated by computing odds ratios (ORs) and 95% confidence intervals (CIs) from the multivariate logistic regression analyses. Genotype analysis and combination analysis using multivariate logistic regression adjusted for age, gender and education levels. A twotailed p-value < 0.05 was considered to indicate statistical significance.
Results
▪ Selection of SNPs in SUMO3
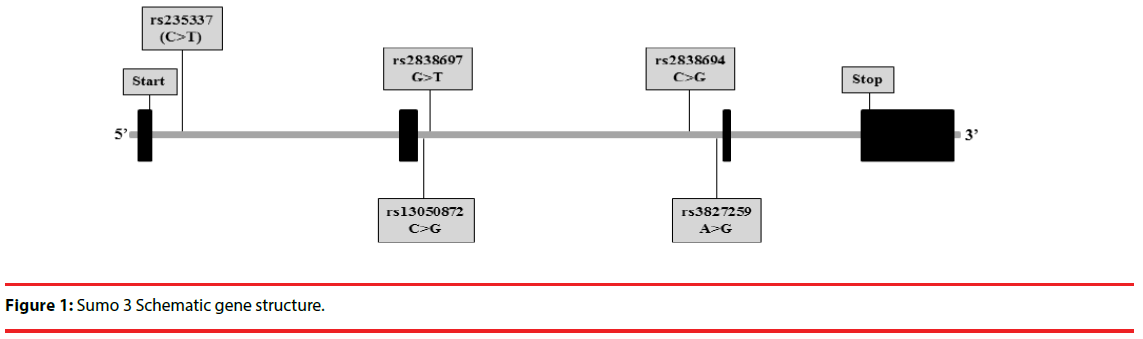
Based on the LD information, minor allele frequency distributions of the five tag SNPs within the SUMO3 gene were 0.0687 for rs23533, 0.2492 for rs3827259, 0.0349 for rs2838694, 0.1460 for rs2838697, and 0.3035 for rs13050872. In addition, the LD information showed that all tag SNPs formed a block of LD with several polymorphisms; however, only rs3827259 polymorphism has not formed the LD block with other polymorphisms. All of the five tag SNPs were detected in the intron region of SUMO3 gene which did not result in a change in amino acid (Figure 1).
▪ Genotype analysis
Genotype frequencies of control group did not deviate from HWE. Our logistic regression analysis results showed that the CT genotype of rs235337 was significantly associated with a reduced risk of AD in a Korean population (OR: 0.448, 95% CI: 0.227-0.885, p-value: 0.021). In contrast, the GG genotype of rs2838694 was significantly associated with a highly increased risk of AD (OR: 34.89, 95% CI: 1.567-776.8, p-value: 0.025) (Table 2). By conducting subgroup analysis of the five tag SNPs retrieved from LD using ApoE ε4 status, the CT genotype of rs235337 was significantly associated with a decreased risk of AD in ApoE ε4 non-carrier group (OR: 0.355, 95% CI: 0.153-0.827, p-value: 0.016). In addition, the GG genotype of rs2838694 polymorphism was related with a highly increased risk of AD in ApoE ε4 noncarrier group (OR: 36.71, 95% CI: 1.579-853.3, p-value: 0.025) (Table 2). However, three other tag SNPs including rs3827259, rs2838697 and rs13050872 did not show association with risk of AD in the ApoE ε4 non-carrier group. In ApoE ε4 carrier status, all tag SNPs were not associated with risk of AD (Table 3). The subgroup analysis by gender showed that the GG genotype of rs2838694 polymorphism was significantly associated with a highly increased risk of AD in female group (OR=26.88, 95% CI=1.197-603.5, p-value=0.038). In contrast, the CT genotype of rs235337 polymorphism was significantly decreased AD risk in female group (OR=0.455, 95% CI=0.211-0.979, p-value=0.044). In male group, the TT genotype of rs235337 polymorphism was significantly associated with highly increased risk of AD (OR=27.80, 95% CI=1.530-541.9, p-value=0.025) (Table 4).
| Case (n) | Control | Adjusted OR (95% CI) a | p-value | HWE in control (p) | ||||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | |||||
| rs235337 (C>T) | CC | 111 | 77.1 | 229 | 70.2 | 1 | ||
| CT | 31 | 21.5 | 90 | 27.6 | 0.448 (0.227-0.885)* | 0.021 | ||
| TT | 2 | 1.4 | 7 | 2.1 | 0.980 (0.131-7.321) | 0.984 | 0.593 | |
| total | 144 | 326 | ||||||
| rs3827259 (A>G) | AA | 131 | 91.0 | 291 | 89.5 | 1 | ||
| AG | 13 | 9.0 | 34 | 10.5 | 0.669 (0.249-1.801) | 0.427 | ||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.320 | |
| total | 144 | 325 | ||||||
| rs2838694 (C>G) | CC | 121 | 84.0 | 244 | 77.2 | 1 | ||
| CG | 22 | 15.3 | 71 | 22.5 | 0.640 (0.304-1.346) | 0.239 | ||
| GG | 1 | 0.7 | 1 | 0.3 | 34.89 (1.567-776.8)* | 0.025 | 0.077 | |
| total | 144 | 316 | ||||||
| rs2838697 (G>T) | GG | 16 | 11.1 | 38 | 12.4 | 1 | ||
| GT | 68 | 47.2 | 158 | 51.6 | 0.727 (0.289-1.827) | 0.498 | ||
| TT | 60 | 41.7 | 110 | 35.9 | 0.912 (0.358-2.325) | 0.847 | 0.103 | |
| total | 144 | 306 | ||||||
| rs13050872 (C>G) | CC | 113 | 78.5 | 248 | 81.0 | 1 | ||
| CG | 31 | 21.5 | 55 | 16.9 | 1.631 (0.818-3.252) | 0.165 | ||
| GG | 0 | 0.0 | 3 | 0.9 | . | . | 0.980 | |
| total | 144 | 306 | ||||||
*statistically significant (p < 0.05)
Table 2: Genotype analysis of five tagSNPs in AD patients and healthy controls.
| AD patients | Healthy controls | Adjusted ORa (95% CI) | p-value | HWE in control (p) | |||||
|---|---|---|---|---|---|---|---|---|---|
| ApoE ε4 non-carrier | n | % | n | % | |||||
| rs235337 (C>T) | CC | 74 | 79.6 | 186 | 69.4 | 1 | |||
| CT | 17 | 18.3 | 77 | 28.7 | 0.355 (0.153-0.827)* | 0.016 | |||
| TT | 2 | 2.2 | 5 | 1.9 | 2.729 (0.306-24.31) | 0.368 | 0.355 | ||
| total | 93 | 268 | |||||||
| rs3827259 (A>G) | AA | 85 | 91.4 | 240 | 89.9 | 1 | |||
| AG | 8 | 8.6 | 27 | 10.1 | 0.791 (0.241-2.592) | 0.699 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.384 | ||
| total | 93 | 267 | |||||||
| rs2838694 (C>G) | CC | 78 | 83.9 | 194 | 75.2 | 1 | |||
| CG | 14 | 15.1 | 63 | 24.4 | 0.537 (0.219-1.319) | 0.175 | |||
| GG | 1 | 1.1 | 1 | 0.4 | 36.71 (1.579-853.3)* | 0.025 | 0.080 | ||
| total | 93 | 258 | |||||||
| rs2838697 (G>T) | GG | 9 | 9.7 | 34 | 13.6 | 1 | |||
| GT | 45 | 48.4 | 131 | 52.4 | 0.903 (0.288-2.833) | 0.861 | |||
| TT | 39 | 41.9 | 85 | 34.0 | 1.051 (0.329-3.360) | 0.933 | 0.139 | ||
| total | 93 | 250 | |||||||
| rs13050872 (C>G) | CC | 75 | 80.6 | 202 | 80.8 | 1 | |||
| CG | 18 | 19.4 | 46 | 18.4 | 1.366 (0.590-3.162) | 0.466 | |||
| GG | 0 | 0.0 | 2 | 0.8 | . | . | 0.725 | ||
| total | 93 | 250 | |||||||
| ApoE ε4 carrier | rs235337 (C>T) | CC | 37 | 72.5 | 43 | 74.1 | 1 | ||
| CT | 14 | 27.5 | 13 | 22.4 | 0.368 (0.078-1.741) | 0.207 | |||
| TT | 0 | 0.0 | 2 | 3.4 | . | . | 0.428 | ||
| total | 51 | 58 | |||||||
| rs3827259 (A>G) | AA | 46 | 90.2 | 51 | 87.9 | 1 | |||
| AG | 5 | 9.8 | 7 | 12.1 | 0.321 (0.040-2.603) | 0.288 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.625 | ||
| total | 51 | 58 | |||||||
| rs2838694 (C>G) | CC | 43 | 84.3 | 50 | 86.2 | 1 | |||
| CG | 8 | 15.7 | 8 | 13.8 | 0.973 (0.210-4.511) | 0.972 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.573 | ||
| total | 51 | 58 | |||||||
| rs2838697 (G>T) | GG | 7 | 13.7 | 4 | 7.1 | 1 | |||
| GT | 23 | 45.1 | 27 | 48.2 | 0.521 (0.078-3.469) | 0.500 | |||
| TT | 21 | 41.2 | 25 | 44.6 | 1.064 (0.153-7.422) | 0.950 | 0.361 | ||
| total | 51 | 56 | |||||||
| rs13050872 (C>G) | CC | 38 | 74.5 | 46 | 82.1 | 1 | |||
| CG | 13 | 25.5 | 9 | 16.1 | 2.278 (0.577-8.995) | 0.240 | |||
| GG | 0 | 0.0 | 1 | 1.8 | . | . | 0.488 | ||
| total | 51 | 56 | |||||||
*statistically significant (p < 0.05)
Table 3: Subgroups analysis of five tagSNPs according to ApoE ε4 status in AD patients and healthy controls.
| AD patients | Healthy controls | Adjusted ORa (95% CI) | p-value | HWE in control (p) | |||||
|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | ||||||
| rs235337 (C>T) | Male | CC | 20 | 80.0 | 116 | 70.3 | 1 | ||
| CT | 4 | 16.0 | 46 | 27.9 | 0.423 (0.094-1.899) | 0.262 | |||
| TT | 1 | 4.0 | 3 | 1.8 | 27.80 (1.530-541.9)* | 0.025 | 0.520 | ||
| total | 25 | 165 | |||||||
| Female | CC | 91 | 76.5 | 113 | 70.2 | 1 | |||
| CT | 27 | 22.7 | 44 | 27.3 | 0.455 (0.211-0.979)* | 0.044 | |||
| TT | 1 | 0.8 | 4 | 2.5 | 0.316 (0.027-3.758) | 0.362 | 0.908 | ||
| total | 119 | 161 | |||||||
| rs3827259 (A>G) | Male | AA | 24 | 96.0 | 145 | 88.4 | 1 | ||
| AG | 1 | 4.0 | 19 | 11.6 | 0.210 (0.013-3.381) | 0.271 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.431 | ||
| total | 25 | 164 | |||||||
| Female | AA | 107 | 89.9 | 146 | 90.7 | 1 | |||
| AG | 12 | 10.1 | 15 | 9.3 | 0.849 (0.286-2.521) | 0.769 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.535 | ||
| total | 119 | 161 | |||||||
| rs2838694 (C>G) | Male | CC | 21 | 84.0 | 124 | 78.5 | 1 | ||
| CG | 4 | 16.0 | 34 | 21.5 | 0.709 (0.128-3.916) | 0.694 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.130 | ||
| total | 25 | 158 | |||||||
| Female | CC | 100 | 84.0 | 120 | 75.9 | 1 | |||
| CG | 18 | 15.1 | 37 | 23.4 | 0.605 (0.266-1.376) | 0.230 | |||
| GG | 1 | 0.8 | 1 | 0.6 | 26.88 (1.197-603.5)* | 0.038 | 0.301 | ||
| total | 119 | 158 | |||||||
| rs2838697 (G>T) | Male | GG | 3 | 12.0 | 22 | 14.4 | 1 | ||
| GT | 10 | 40.0 | 75 | 49.0 | 0.749 (0.123-4.565) | 0.754 | |||
| TT | 12 | 48.0 | 56 | 36.6 | 0.603 (0.097-3.762) | 0.588 | 0.698 | ||
| total | 25 | 153 | |||||||
| Female | GG | 13 | 10.9 | 16 | 10.5 | 1 | |||
| GT | 58 | 48.7 | 83 | 54.2 | 0.750 (0.257-2.187) | 0.598 | |||
| TT | 48 | 40.3 | 54 | 35.3 | 1.048 (0.351-3.134) | 0.932 | 0.053 | ||
| total | 119 | 153 | |||||||
| rs1305872 (C>G) | Male | CC | 20 | 80.0 | 120 | 78.4 | 1 | ||
| CG | 5 | 20.0 | 30 | 19.6 | 1.816 (0.433-7.616) | 0.415 | |||
| GG | 0 | 0.0 | 3 | 2.0 | . | . | 0.492 | ||
| total | 25 | 153 | |||||||
| Female | CC | 93 | 78.2 | 128 | 83.7 | 1 | |||
| CG | 26 | 21.8 | 25 | 16.3 | 1.604 (0.729-3.528) | 0.240 | |||
| GG | 0 | 0.0 | 0 | 0.0 | . | . | 0.271 | ||
| total | 119 | 153 | |||||||
*statistically significant (p<0.05)
Table 4: Subgroups analysis of five tagSNPs according to gender in AD patients and healthy controls.
Discussion
Until recently, few genetic studies of the association between sumoylation related genes polymorphisms and risk of AD have been reported. In 2007, Grupe et al. investigated the susceptibility genes for late-onset AD (LOAD) in UK and USA large subjects using a genome wide association study and identified the 19 LOAD susceptible polymorphisms including rs6907175 of LOC651924. Interestingly, LOC651924 is homolog of SUMO1 activating enzyme subunit 2 [20]. Two years later, Ahn et al. reported that the rs761059 polymorphism of the UBC9 gene was significantly associated with an increased risk of AD in a Korean population. They reported that the two polymorphisms including rs8052688 and rs8063 were significantly associated with a risk of mild cognitive impairment [21]. Our research group has studied the association between SUMO genes polymorphisms and risk of AD in a Korean population and demonstrated that three polymorphisms including rs12472035, rs35271045 and rs9913676 of SUMO1 and SUMO2 genes were significantly associated with an increased risk of AD in a Korean population [18]. Several studies have focused on the association with SUMO proteins and pathogenesis of neurodegenerative disease; however, there have not been a single study focused on the genetic association between SUMO3 and risk of AD.
In our case-control study on SUMO3, three risk factors of AD including age, gender and education levels were adjusted for data analysis. Age is greatest important risk factor of AD and many elderly people over 65 years of age are occurred AD [1]. In addition, the prevalence and incidence rates of AD showed significant difference according to gender, and female group is significantly related with high risk of AD pathogenesis [22,23], and education level was significantly associated with AD pathogenesis [24]. It has been reported that risk of developing AD was significantly decreased in patients with 8-9 years of education compared to patients with 6-7 years of education [24]. Therefore, our statistical analysis has adjusted by age, gender and education levels. In this present study, we observed that the rs235337 polymorphism was significantly associated with an increased risk of AD, whereas the rs2838694 polymorphism was significantly associated with a decreased risk of AD. In subgroup analysis by gender, the GG genotype of rs2838694 polymorphism was significantly associated with a highly increased risk of AD in female group. In contrast, the CT genotype of rs235337 polymorphism was significantly decreased AD risk in female group. In male group, the TT genotype of rs235337 polymorphism was significantly associated with a highly increased risk of AD. Based on these results, we suggest that rs235337 and rs23838694 polymorphisms may be associated with risk of AD in female group. Out study for the first time showed a link between the SUMO3 polymorphisms and AD risk using a population.
Past decades, many studies were demonstrated that ApoE ε4 carrier has been well-established as a susceptible gene. The ApoE gene located in the chromosome 19q13.32 has three major alleles including ε2, ε3 and ε4, and the association between ApoE ε4 carrier and prevalence of AD using comprehensive meta-analysis was examined [25]. This meta-analysis demonstrated that ApoE heterozygote (ε4/–) genotype caused high prevalence of probable AD in many ethnicity excluding South America. Similarly, ApoE homozygote (ε4/ε4) genotype was also significantly associated with highly increased prevalence of probable AD in worldwide. The statistical analysis showed that heterozygote genotype of ApoE ε4 carrier was associated with increased prevalence of AD in many countries including Australia, China, Colombia, Finland, Germany, Italy, Japan, Netherlands, Sweden, UK and USA, while no association was observed in Canada, France, Spain and South Korea [25]. Although these results of ApoE ε4 carrier were controversial by country or ethnicity, a number of studies indicated that ApoE ε4 carrier was significantly associated with AD; therefore, our data were stratified by the ApoE ε4 status.
The current case-control study has several limitations. First, our sample size is small. Small sample size may be affected to reduction of statistical power. Therefore, our case-control study needs to re-examine using large-sample size. Second, risk factors which affect to AD are greatly various. However, we have investigated only few risk factors including age, gender, education levels and ApoE status in our casecontrol study. Further study is considered various environmental factors including alcohol consume, smoking status, social abilities and others. In addition, we also considered many genetic risk factors. Interaction between genes may be help to identification of specific genetic risk factors for AD. Finally, we need to consider results of clinical test in our case-control study. Many psychologists have been investigated severity of dementia using various tests including mini–mental state examination (MMSE), clinical dementia rating (CDR) and other tests. Fusion research of the association between genetic polymorphisms and clinical test may be provided the interesting results. Despite the limitations, our case-control study provided the evidences of the association between SUMO3 polymorphisms and risk of AD. The rs235337 and rs2838694 SUMO polymorphisms were statistically associated with the risk of AD in Korean population. Although future functional and replication studies are needed to elucidate the role of SUMO3 in AD for determining the association between the SUMO3 polymorphisms and AD, our results show that two polymorphisms in SUMO3 are involved in the pathogenesis of AD and could be potential genetic risk factors for AD.
Conflict of Interest Statement
The authors declare no conflict of interests
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant number: 2009-0093829).
References
- Alzheimer's A. 2016 Alzheimer's disease facts and figures. Alzheimer's & Dementia12(4), 459-509 (2016).
- Organization WH. Dementia; a public health priority (2012).
- Gendron TF, Petrucelli L. The role of tau in neurodegeneration. Molecular. neurodegeneration 4(1), 13 (2009).
- Zhang YW, Thompson R, Zhang H, et al. APP processing in Alzheimer's disease. Molecular. Brain 4(1), 3 (2011).
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer's disease and related disorders. Nature. Reviews. Neuroscience 8(9), 663-672 (2007).
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nature. Reviews. Molecular Cell Biology 8(12), 947-956 (2007).
- Melchior F. SUMO--nonclassical ubiquitin. Annual. Review. Of. cell. And. Developmental. biology 16(1), 591-626 (2000).
- Bohren KM, Gabbay KH, Owerbach D. Affinity chromatography of native SUMO proteins using His-tagged recombinant UBC9 bound to Co2+-charged talon resin. Protein. Expression. And .purification 54(2), 289-294 (2007).
- Mo YY, Yu Y, Theodosiou E, et al. A role for Ubc9 in tumorigenesis. Oncogene 24(16), 2677-2683 (2005).
- Cheng J, Bawa T, Lee P, et al. Role of desumoylation in the development of prostate cancer. Neoplasia 8(8), 667-676 (2006).
- Sarge KD, Park-Sarge OK. Sumoylation and human disease pathogenesis. Trends. In. Biochemical. Sciences 34(4), 200-205 (2009).
- Li Y, Wang H, Wang S, et al. Positive and negative regulation of APP amyloidogenesis by sumoylation. Procee. Of. the. National. Aca. Of. Sciences. Of. the. United. States. Of. America 100(1), 259-264 (2003).
- Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochimica. Et. Biophysica. Acta 1773(6), 694-706 (2007).
- Jacobsen JS, Wu CC, Redwine JM, et al. Early-onset behavioral and synaptic deficits in a mouse model of Alzheimer's disease. Procee. Of. the. National. Aca. Of. Sciences. Of. the. United. States. Of. America 103(13), 5161-5166 (2006).
- Kim TH, Jhoo JH, Park JH, et al. Korean version of mini mental status examination for dementia screening and its' short form. Psychiatry. Investigation 7(2), 102-108 (2010).
- Lee JH, Lee KU, Lee DY, et al. Development of the Korean version of the Consortium to Establish a Registry for Alzheimer's Disease Assessment Packet (CERAD-K): clinical and neuropsychological assessment batteries. Journal of Gerontol 57(1), P47-P53 (2002).
- McKhann G, Drachman D, Folstein M, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34(7), 939-944 (1984).
- Mun MJ, Kim JH, Choi JY, et al. Polymorphisms of small ubiquitin-related modifier genes are associated with risk of Alzheimer's disease in Korean: A case-control study. Journal. Of. the. Neurological. Sciences 364(1), 122-127 (2016).
- Lee DY, Lee KU, Lee JH, et al. A normative study of the CERAD neuropsychological assessment battery in the Korean elderly. JINS 10(1), 72-81 (2004).
- Grupe A, Abraham R, Li Y, et al. Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Human Molecular Genetics 16(8), 865-873 (2007).
- Ahn K, Song JH, Kim DK, et al. Ubc9 gene polymorphisms and late-onset Alzheimer's disease in the Korean population: a genetic association study. Neuroscience Letters 465(3), 272-275 (2009).
- Di Carlo A, Baldereschi M, Amaducci L, et al. Incidence of dementia, Alzheimer's disease, and vascular dementia in Italy. The ILSA Study. Journal. Of. the. American. Geriatrics. Society 50(1), 41-48 (2002).
- Andersen K, Launer LJ, Dewey ME, = et al. Gender differences in the incidence of AD and vascular dementia: The EURODEM Studies. EURODEM Incidence Research Group. Neurology 53(9), 1992-1997 (1999).
- Sando SB, Melquist S, Cannon A, et al. Risk-reducing effect of education in Alzheimer's disease. International. Journal. Of. geriatric. psychiatry 23(11), 1156-1162 (2008).
- Ward A, Crean S, Mercaldi CJ, et al. Prevalence of apolipoprotein E4 genotype and homozygotes (APOE e4/4) among patients diagnosed with Alzheimer's disease: a systematic review and meta-analysis. Neuroepidemiology 38(1),1-17 (2012).