Research Article - (2018) Volume 8, Issue 4
Altered Cd8+ T lymphocyte Response Triggered by Arginase 1: Implication for Fatigue Intensification during Localized Radiation Therapy in Prostate Cancer Patients
- Corresponding Author:
- Xiao-Min Wang
Department of Anesthesiology, the University of Hong Kong, Hong Kong
Tel: +81-3-3819-7270
Abstract
Fatigue, the most common side effect of cancer treatments, is observed to intensify during external-beam radiation therapy (EBRT). The underlying molecular mechanisms remain unclear. This study investigated the differentially expressed genes/proteins and their association with fatigue intensification during EBRT. Fatigue scores measured by FACT-F and peripheral blood were collected prior to treatment (baseline D0), at midpoint (days 19-21, D21) and endpoint (days 38-42, D42) from men (n=30) with non-metastatic prostate cancer undergoing EBRT. RNA extracted from peripheral blood was used for gene expression analysis. Plasma arginase I and arginine were examined using ELISA and liquid chromatography–tandem mass spectrometry. Differences in fatigue scores, gene and protein expression between times points following EBRT were analyzed by one way ANOVA followed by Post Hoc t-test. Fatigue scores decreased significantly from baseline (44.6 ± 8.1) to midpoint (37.3 ± 10.6, p=0.000, low scores indicating high fatigue) and to endpoint (37.4 ± 10.1, p=0.001) during EBRT. ARG1 (encoding arginase type 1) was significantly up regulated from baseline to midpoint of EBRT (fold change =2.41, p<0.05) whereas genes associated with the adaptive immune functional pathway (CD28, CD27, CCR7, CD3D, CD8A and HLA-DOB) were significantly downregulated >2-fold between the study time points. The changes in gene expression were associated with patient reported fatigue intensity. Moreover, the upregulation of ARG1 was negatively correlated with the absolute lymphocyte count (R2=0.561, p=0.01) only in the high level of fatigue group (n=17) during EBRT. Increased ARG1 expression is known to result in arginine deficiency, which leads to immunosuppression by impairing lymphocyte proliferation and activation. EBRT-induced ARG1 upregulation may play an essential role in fatigue intensification via the arginine deficiency and suppression of T-cell proliferation pathways. These findings may provide novel insights into the molecular-genetic mechanisms underlying the development and intensification of cancer treatment-related fatigue.
Keywords
Cancer-related fatigue (CRF), Prostate cancer, External beam radiation therapy (EBRT), Lymphocyte, Gene expression
Background
Cancer related fatigue (CRF) is defined as a distressing and persistent subjective feeling of tiredness and exhaustion that is not proportionate to the level of activity or energy exerted, nor completely relieved by sleep and rest [1-4]. It is the most common side effect of cancer treatment including chemotherapy, radiation therapy (RT), or selected biologic response modifiers [5]. Cancer related fatigue affects more than 80% of cancer patients during treatment [6], and nearly 40% of cancer survivors [7], which negatively impacts the health-related quality of life (QOL) of these individuals [8]. Four percent of these patients experienced severe fatigue up to 5 years post cancer treatment [9].
RT for non-metastatic cancer, especially with the use of modulated radiation therapy such as external beam radiation therapy (EBRT) has increased cure and survival rates of patients [10]. RT can however result in numerous side effects, specifically side effects related to targeted organs, including urinary, bowel, and sexual dysfunctions, as well as fatigue in men with non-metastatic prostate cancer receiving EBRT in the pelvic area. [11]. In fact, 71% of these men report clinically significant fatigue while receiving RT [12]. Unlike fatigue related to chemotherapy, which reaches a peak in the days following treatment and then decreases until the next treatment, the fatigue induced by RT, even peripherally-targeted RT often increases gradually in intensity over the course of treatment [13]. There is no U.S. Food and Drug Administration (FDA) approved pharmacological intervention for CRF. During active treatment, the most current National Comprehensive Cancer Network (NCCN) clinical practice guidelines recommended the use of methylphenidate (psychostimulants) for the pharmacological intervention for cancer related fatigue [14]. However, a systematic review and meta-analysis reported the inconsistent effectiveness of methylphenidate on CRF [15]. One possible explanation of this inconsistency and ineffectiveness of treatment for CRF may be due to lack of complete understanding of CRF etiology and mechanisms underlying its development.
Few studies have reported the associations of CRF with impairment in immune function, especially related to T lymphocyte activities. However, the results are inconsistent. In one study, fatigued breast cancer survivors had significantly higher circulating T-cell subsets (CD3+ T lymphocytes, CD4+ T lymphocytes, and CD3+/CD56+ effector T lymphocytes) than non-fatigued breast cancer survivors [16]. Another study conducted by the same research group found that fatigued breast cancer survivors had lower levels of circulating effector T-cells (CD3+/CD69+) [17]. Although these two studies suggested the role of T-cell mediated inflammation in the development of CRF via cytokine signaling pathways [16,17], further study exploring the association of T-cell alteration, inflammation, and the development of CRF is needed. Particularly, defining the molecular-genetic mechanisms underlying the initiation, intensification, and progression of CRF is critical to developing efficient preventive and treatment strategies. Not only are these strategies necessary to enhance functional status and health-related QOL of patients during cancer therapy, but also during survivorship. Therefore, this study investigated the differential expression of genes and proteins associated with T lymphocyte proliferation, function and their reciprocal associations with self-reported fatigue intensification in men receiving EBRT for nonmetastatic prostate cancer.
Materials and Methods
▪ Subjects, timeline of clinical procedures and sample collection
Subjects with non-metastatic prostate cancer scheduled to receive EBRT were enrolled under an actively recruiting protocol NCT01143467. The protocol was approved by the Institutional Review Board of the National Institutes of Health (NIH), Bethesda, Maryland, USA. Recruitment and data collection were conducted from May 2009 to December 2012 at the Hatfield Clinical Research Center, NIH. Informed consent was obtained from all participants. Patients were excluded if they had any progressive disease causing fatigue; psychiatric disorder within five years; uncorrected hypothyroidism or anemia; second malignancies; or were taking sedatives, steroids, or non-steroidal anti-inflammatory agents.
Fatigue and peripheral blood were collected at baseline (prior to EBRT, D0), midpoint (days 19–21, D21) and endpoint (days 38-42, D 42) of EBRT, when fatigue intensification gradually increased in our previous study [2]. Fatigue intensity was measured using the Functional Assessment of Cancer Therapy-Fatigue subscale (FACT-F), a 13-item self-report questionnaire. The FACT-F is a frequently used, validated, reliable, and stand-alone measure of fatigue in cancer therapy with coefficient alphas greater than 90 [18,19]. The scores on the instrument range from 0-52, with higher scores indicating less fatigue. In the general population, the mean FACT-F score was 43.6 ± 9.4, whereas in nonanemic cancer patients, it was 40.0 ± 9.8 [18]. A change of 3-point or higher in FACT-F scores is commonly accepted as representing clinically significant and meaningful change in fatigue [19]. All participants were also evaluated using the Hamilton Depression Rating Scale (HAM-D) to exclude subjects with depression. The HAM-D is a 21-item, clinician-rated paper questionnaire with good internal reliability (α=0.81 to 0.98) [20].
Peripheral blood was collected from each subject at each time point to obtain a complete blood count (CBC) and RNA gene analysis using EDTA and PAXgene blood RNA tubes (Qiagen, Frederick, MD, USA), respectively. CBC was analyzed by the NIH Department of Laboratory Medicine using standard hospital procedure. All other samples were stored at -80oC until RNA extraction was performed to reduce intraassay variability. Total RNA was isolated per manufacturer’s protocol. RNA concentration, purity, and integrity were determined using the NanoDrop ((ND-1000; Wilmington, Delaware) and Experion systems (BioRad, Hercules, California), respectively.
▪ Gene expression analysis using Affymetrix microarray
In the study, 90 GeneChips Human Genome U133 Plus 2.0 were used (n=10 at each time point following EBRT). All microarray chips were processed in the same laboratory following standard operating protocols to minimize nonbiological technical bias. A total of 100 ng of purified RNA was used to reverse-transcribe into cDNA, which was used for the synthesis of biotin-labeled cRNA. Subsequently, 15 μg of fragmented cRNA was used to hybridize each chip at 45°C for 16 hours in a GeneChip hybridization oven at 60 rpm. Affymetrix GeneChip Command Console (AGCC, 3.0 V) was used to scan the images and gather raw data. Microarray quality control (QC) was evaluated for each Affymetrix array. Raw data (Affymetrix. CEL files) were imported into Partek® Genomic SuiteTM (Partek Inc., St. Louis, MO). Probe summarization and probe-set normalization were performed using Robust Multi-Chip Average (RMA), which includes background correction, quartile normalization, log2 transformation, and median polish probe set summarization. Principal Component Analysis (PCA) was used to detect outliers. Differences in gene expression between study time points during EBRT were analyzed using repeated measures multiway ANOVA followed by post hoc pairwise comparison to determine the differential gene expression at different time points during EBRT. Differentially expressed genes were identified by at least a 2-fold change in either direction with the p value corrected for false discovery rate (FDR of 0.05). Batch effects were controlled in the analysis by including the scanned dates in the ANOVA model. Biological pathway analysis was performed using Pathway Enrichment.
▪ Verification by quantitative real-time PCR (qRT-PCR)
Differentially expressed genes that were significantly correlated at r>0.50 and p<0.05 with FACT-F scores were selected for validation using the same RNA samples used in the microarray analysis. Quantitative real time PCR (qRT-PCR) was performed by the Laboratory of Molecular Technology at Frederick National Laboratory for Cancer Research (SAICFrederick, Maryland). All reagents used in qRTPCR were purchased from Applied Biosystems (Foster City, CA). Briefly, 50 ng of RNA in 5 ul of DNase-treated RNA plus 5 μl master mix in 10 μl reaction was used to synthesize cDNA using an ABI High Capacity cDNA reverse transcription kit (ABI 4374967). PCR was performed with Pool TaqMan assays from 20X to 0.2X, including two other genes RNase P and GAPDH. Complementary DNA template was synthesized using ABI PreAmp master mix (4391128): 5.0 μl 2X master mix + 2.5 μl pooled assays + 2.5 μl RT products, 15 cycles. DNA suspension buffer (20 μl) was added to each reaction and used 2 μl for 5 μl TaqMan reactions with GAPDH and RNase P. Ct ranged from 7-15. Quantification of gene expression was performed using Fluidigm 96.96 dynamic gene expression array. Data were exported and compiled with Excel.
▪ Plasma arginase I and arginine detection
Concentrations of arginase I and arginine were examined using plasma collected in EDTA tubes from the same subjects in the microarray analysis at all study time points. The plasma samples were separated from whole blood and stored at -80°C. Frozen plasma samples were thawed on ice and centrifuged at 1,000 xg for ten minutes at 4°C. ELISA was performed using 20-50 μl of nonediluted plasma samples. Protein confirmation was tested in triplicate. Each plate was read by VICTOR3 at 450 nm. Arginase I was measured by ELISA (Hycult Biotech, Plymouth Meeting, PA) following the manufacturer’s instructions.
Arginine in the plasma was extracted by protein precipitation with 5%TCA containing 20 uM 13C6-arginine (internal standard). The supernatant was analyzed by liquid chromatography – tandem mass spectrometry (LC-MS/MS) with electrospray ionization. Arginine concentrations in the samples were determined from the linear regression curve (1-250 uM) constructed by plotting the peak area ratios against the standard concentrations. The reference limits for plasma L-arginine were established, using a stable isotope based LC-MS/ MS method, at the 2.5th and 97.5th percentiles to be 41.0 μmol/L (95% CI=39.5-42.5 μmol/L) and 114 μmol/L (95% CI=112-115 μmol/L), respectively [21].
▪ Data analyses
As previously described [2,22], descriptive statistics were performed to identify the participants’ demographic characteristics. The differences in fatigue intensity, gene and protein expressions at different time points following EBRT were analyzed using one way repeated measures ANOVA followed by post-hoc t-test to compare the differences between control at baseline (D0) to midpoint (D21) and endpoint (D42) during EBRT. The association among the gene expression of interest and the association between the gene expression and patient reported fatigue scores were examined using Pearson correlation. All statistical analyses were conducted using SPSS software version 21 (IBM Corporation, Armonk, New York).
Results
▪ Study population, demographics and fatigue scores
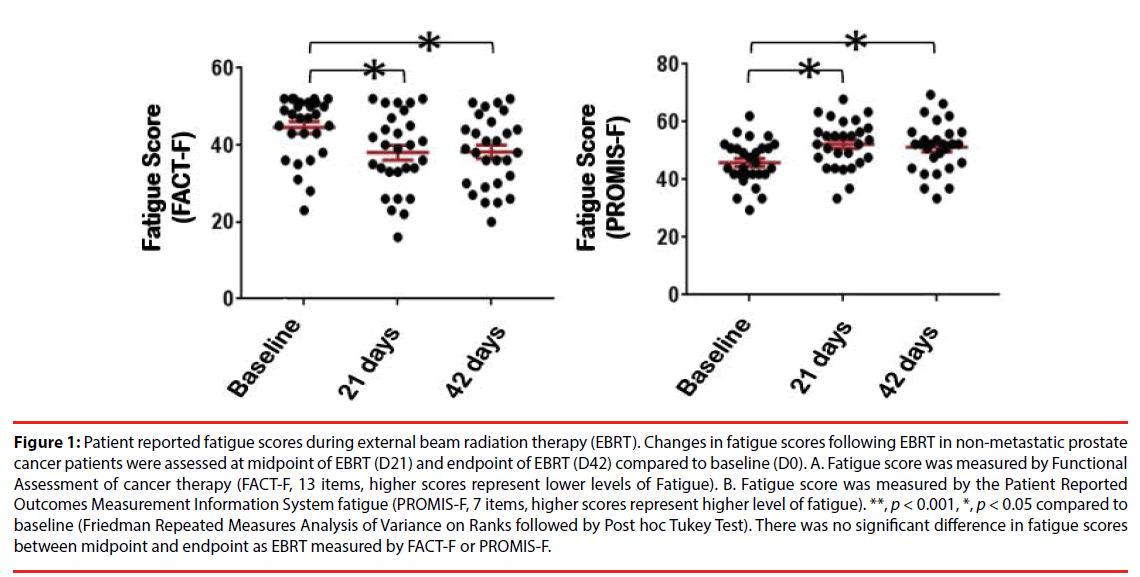
A total of 30 men with non-metastatic prostate cancer (73% Caucasian, 23% African American and 3% other race) between 49-81 years old were included in this study. The demographic and clinical characteristics of the sample are shown in (Table 1). None of the participants reached the cutoff score for depression as measured by the HAM-D either at baseline (mean=1.13 ± 1.76, range 0-8) or at midpoint of EBRT (mean=2.17 ± 3.38, range 0-13) or endpoint (mean=1.37 ± 1.60, range 0-8). Depression levels in the three time points were not statistically significant. Of the 30 subjects, 87% (26/30) received a total dose of 75.6 Gray with EBRT, while the remaining 13% (4/30) received a total dose of 68.4 Gray. Fatigue scores, as measured by FACT-F (low scores indicating high fatigue), significantly decreased from baseline (mean=44.5 ± 8.1) to midpoint (mean=37.3 ± 10.6, p=0.000) and from baseline to endpoint (mean=37.4 ± 10.1, p=0.001) following EBRT. There was no significant difference of fatigue level between midpoint and endpoint of EBRT (Figure 1A), p=0.915). To verify the outcomes of FACT-F assessment, fatigue scores were further evaluated using the Patient Reported Outcomes Measurement Information System fatigue (PROMIS-F) scale, 7 items, higher scores represent higher levels of fatigue, (Figure 1B) in the same patients during EBRT. As shown in (Figure 1), patient-reported fatigue intensities were peaked at midpoint (D21) and there was no significant difference in terms of fatigue scores between midpoint and endpoint. These findings indicate that in these non-metastatic prostate cancer patients undergoing EBRT, fatigue developed and its intensity reached a peak level at the midpoint of EBRT and maintained near its peak level at completion of EBRT. Thus, to investigate the differential gene expression and the possible biological pathway underlying fatigue intensification during EBRT, we then focused on comparing the differences between baseline (D0) and midpoint (D21) in the following results and discussion sections.
| Mean | SD | Range | N (%) | Repeated measure |
|---|---|---|---|---|
| ANOVA p value | ||||
| Age in Years | 64.3 | 7.5 | 49-81 | 30 (100) |
| Race | ||||
| Caucasian | 22 (73) | |||
| African-American | 7 (23) | |||
| Other | ||||
| T1 (a-c) | 11 (36.7) | |||
| T2 (a-c) | 17 (56.7) | |||
| T3 (a-c) | 2 (6.7) | |||
| Gleason score | 7.6 | 0.99 | 6-9 | |
| BMI | 29.5 | 4.1 | 22.9-40.2 | |
| Total Dosage EBRT (Gray) | ||||
| 75.6 | 26 (87) | |||
| 68.4 | 4 (13) | |||
| PSA Levels (ng/mL) | ||||
| Baseline | 10.4 | 10.7 | 0.08-33.4 | |
| Completion | 0.23 | 0.60 | 0.01-2.59 | |
| Albumin levels (g/dL) | ||||
| Baseline | 4.1 | 0.80 | 3.6-4.5 | |
| Testosterone (ng/dL) | ||||
| Baseline | 292.6 | 174 | 20-505 | |
| TSH (mcIU/ml) | ||||
| Baseline | 2.26 | 1.44 | 0.24-4.15 | |
| Hemoglobin | ||||
| Baseline | 13.5 | 1.10 | 10.9-15.1 | |
| FACT-F Questionnaire Baseline | 44.57 | 8.1 | 23-52 | |
| Midpoint | 37.34 | 10.6 | 16-52 | 0.000a |
| Endpoint | 37.38 | 10.1 | 14-52 | 0.001 |
| HAM-D Questionnaire | ||||
| Baseline | 1.13 | 1.76 | 0-8 | |
| Midpoint | 2.17 | 3.38 | 0-13 | 0.137 |
| Endpoint | 1.37 | 1.67 | 0-8 | |
Note:aRepeated measure p value <0.01, Pairwise comparisons showed significant difference in mean between baseline and midpoint of EBRT (p=0.000), and between baseline and endpoint of EBRT (p=0.001). There is no significant difference in mean between midpoint and endpoint of EBRT (p=0.915).
Table 1: Demographics and Clinical Characteristics of Sample (N=30).
Figure 1: Patient reported fatigue scores during external beam radiation therapy (EBRT). Changes in fatigue scores following EBRT in non-metastatic prostate cancer patients were assessed at midpoint of EBRT (D21) and endpoint of EBRT (D42) compared to baseline (D0). A. Fatigue score was measured by Functional Assessment of cancer therapy (FACT-F, 13 items, higher scores represent lower levels of Fatigue). B. Fatigue score was measured by the Patient Reported Outcomes Measurement Information System fatigue (PROMIS-F, 7 items, higher scores represent higher level of fatigue). **, p < 0.001, *, p < 0.05 compared to baseline (Friedman Repeated Measures Analysis of Variance on Ranks followed by Post hoc Tukey Test). There was no significant difference in fatigue scores between midpoint and endpoint as EBRT measured by FACT-F or PROMIS-F.
▪ Gene expression profile and regulatory networks of fatigue intensification
In order to clearly describe the changes in gene expression at the initial fatigue intensification during EBRT, the fold changes in gene expression where compared from D0 prior to EBRT to D21 following EBRT were acquired by use of microarray gene analysis. Compared to D0, there were 327 transcripts with over a 2-fold change in expression at D21 (FDR adjusted p<0.05, n=30). The top 10 upregulated and downregulated genes are listed in Table 2. The differentially expressed genes were then analyzed for their associations with canonical pathways. The top three canonical pathways were the T-cell receptor signaling (p=3.48 × 1012), the calciuminduced T lymphocyte apoptosis (p=1.42 × 1011), and the iCOS-iCOSL signaling in T helper cells (p=8.35 × 1011) (Table 3).
| Probeset ID | Gene symbol | Fold change | P value | Gene Title |
|---|---|---|---|---|
| Top 10 up regulated genes | ||||
| 205950_s_at | CA1 | 3.33 | 1.22 × 107 | carbonic anhydrase I |
| 206698_at | XK | 2.62 | 2.09 × 105 | X-linked Kx blood group (McLeod syndrome) |
| 236081_at | SNCA | 2.60 | 1.04 × 105 | synuclein, alpha (non A4 component of amyloid precursor) |
| 223669_at | HEMGN | 2.46 | 6.63 × 106 | ATP-binding cassette, sub-family C (CFTR/MRP), member 13 |
| 214407_x_at | GYPB | 2.43 | 8.62 × 107 | glycophorin B (MNS blood group) |
| 206177_s_at | ARG1 | 2.41 | 3.44 × 105 | arginase, liver |
| 203502_at | BPGM | 2.31 | 5.56 × 105 | 2,3-bisphosphoglycerate mutase |
| 226811_at | FAM46C | 2.28 | 5.27 × 105 | family with sequence similarity 46, member C |
| 203116_s_at | FECH | 2.17 | 6.54 × 105 | ferrochelatase |
| 1552583_s_at | ABCC13 | 2.16 | 8.54 × 108 | ATP-binding cassette, sub-family C (CFTR/MRP), member 13, pseudogene |
| Top 10 down regulated genes | ||||
| 228592_at | MS4A1 | -8.53 | 1.36 × 1016 | membrane-spanning 4-domains, subfamily A, member 1 |
| 212827_at | IGHM | -6.83 | 3.81 × 1019 | immunoglobulin heavy constant mu |
| 221969_at | PAX5 | -4.32 | 1.51 × 1017 | paired box 5 |
| 206337_at | CCR7 | -4.25 | 4.73 × 1014 | chemokine (C-C motif) receptor 7 |
| 205267_at | POU2AF1 | -4.24 | 7.21 × 1016 | POU class 2 associating factor 1 |
| 236796_at | BACH2 | -3.90 | 5.88 × 1018 | B-cell scaffold protein with ankyrin repeats 1 |
| 230245_s_at | LOC283663 | -3.85 | 1.98 × 1018 | hypothetical LOC283663 |
| 39318_at | TCL1A | -3.50 | 2.00 × 1016 | T-cell leukemia/lymphoma 1A |
| 231093_at | FCRL3 | -3.45 | 2.32 × 1012 | Fc receptor-like 3 |
| 229513_at | STRBP | -3.32 | 7.28 × 1018 | spermatid perinuclear RNA binding protein |
Table 2: Top differentially expressed genes between baseline and midpoint of EBRT (N = 30).
| Symbol | Gene name | Probeset ID | p-value | Fold Change | |||
|---|---|---|---|---|---|---|---|
| T cell Receptor signaling (p=3.48x1012, Ratio=14/109 (0.128) | |||||||
| CAMK4 | calcium/calmodulin-dependent protein kinase IV | 229029_at | 5.09E-10 | -2.558 | |||
| CD28 | CD28 molecule | 206545_at | 1.56E-06 | -2.111 | |||
| CD247 | CD247 molecule | 210031_at | 1.66E-08 | -2.330 | |||
| CD3D | CD3d molecule, delta (CD3-TCR complex) | 213539_at | 1.58E-06 | -2.057 | |||
| CD3G | CD3g molecule, gamma (CD3-TCR complex) | 206804_at | 2.37E-07 | -2.114 | |||
| CD8A | CD8a molecule | 205758_at | 3.16E-06 | -2.174 | |||
| ITK | IL2-inducible T-cell kinase | 211339_s_at | 9.98E-08 | -2.326 | |||
| LAT | linker for activation of T cells | 211005_at | 2.19E-10 | -2.198 | |||
| LCK | lymphocyte-specific protein tyrosine kinase | 204891_s_at | 7.45E-11 | -2.536 | |||
| NFATC2 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 226991_at | 4.69E-09 | -2.074 | |||
| PRKCQ | protein kinase C, theta | 210038_at | 6.59E-09 | -2.023 | |||
| RASGRP1 | RAS guanyl releasing protein 1 (calcium and DAG-regulated) | 205590_at | 1.49E-06 | -2.372 | |||
| RRAS2 | related RAS viral (r-ras) oncogene homolog 2 | 212589_at | 3.71E-06 | -2.251 | |||
| TXK | TXK tyrosine kinase | 206828_at | 9.63E-09 | -2.510 | |||
| Calcium-induced T Lymphocyte Apoptosis (p=1.42x1011, Ratio=11/71 (0.155) | |||||||
| CAMK4 | calcium/calmodulin-dependent protein kinase IV | 229029_at | 5.09E-10 | -2.558 | |||
| CD247 | CD247 molecule | 210031_at | 1.66E-08 | -2.330 | |||
| CD3D | CD3d molecule, delta (CD3-TCR complex) | 213539_at | 1.58E-06 | -2.057 | |||
| CD3G | CD3g molecule, gamma (CD3-TCR complex) | 206804_at | 2.37E-07 | -2.114 | |||
| HLA-DOB | major histocompatibility complex, class II, DO beta | 205671_s_at | 8.02E-11 | -2.080 | |||
| ITPR1 | inositol 1,4,5-trisphosphate receptor, type 1 | 240052_at | 1.02E-10 | -2.288 | |||
| LCK | lymphocyte-specific protein tyrosine kinase | 204891_s_at | 7.45E-11 | -2.536 | |||
| NFATC2 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 226991_at | 4.69E-09 | -2.074 | |||
| PRKCA | protein kinase C, alpha | 213093_at | 6.22E-08 | -2.316 | |||
| PRKCH | protein kinase C, eta | 218764_at | 6.70E-09 | -2.216 | |||
| PRKCQ | protein kinase C, theta | 210038_at | 6.59E-09 | -2.023 | |||
| iCOS-iCOSL Signaling in T Helper Cells (p=8.35x1011, Ratio=13/126 (0.103) | |||||||
| CAMK4 | calcium/calmodulin-dependent protein kinase IV | 229029_at | 5.09E-10 | -2.558 | |||
| CD28 | CD28 molecule | 206545_at | 1.56E-06 | -2.111 | |||
| CD247 | CD247 molecule | 210031_at | 1.66E-08 | -2.330 | |||
| CD3D | CD3d molecule, delta (CD3-TCR complex) | 213539_at | 1.58E-06 | -2.057 | |||
| CD3G | CD3g molecule, gamma (CD3-TCR complex) | 206804_at | 2.37E-07 | -2.114 | |||
| HLA-DOB | major histocompatibility complex, class II, DO beta | 205671_s_at | 8.02E-11 | -2.080 | |||
| ITK | IL2-inducible T-cell kinase | 211339_s_at | 9.98E-08 | -2.326 | |||
| ITPR1 | inositol 1,4,5-trisphosphate receptor, type 1 | 240052_at | 1.02E-10 | -2.288 | |||
| LAT | linker for activation of T cells | 211005_at | 2.19E-10 | -2.198 | |||
| LCK | lymphocyte-specific protein tyrosine kinase | 204891_s_at | 7.45E-11 | -2.536 | |||
| NFATC2 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 | 226991_at | 4.69E-09 | -2.074 | |||
| PLEKHA1 | pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 | 219024_at | 6.50E-09 | -2.171 | |||
| PRKCQ | protein kinase C, theta | 210038_at | 6.59E-09 | -2.023 | |||
Table 3: Top 3 canonical pathways and genes associated with each pathway.
The specific differentially expressed genes of interest that were associated with the top 3 canonical pathways were the upregulation of ARG1 (encoding arginase type 1, fold change =2.41, adjusted p<0.001), CA1 (carbonic anhydrase 1, fold change =3.33, adjusted p<0.001), and XK (X-linked Kx blood group, fold change =2.62, adjusted p<0.001), as well as the down regulation of CD8A (encoding the CD8 alpha chain, fold change =-2.17, adjusted p<0.001), CD27 (encoding TNF-receptor superfamily, fold change =-2.11, adjusted p<0.001), CD28 (encoding CD28 molecule, fold change =-2.11, adjusted p<0.001), and CCR7 (chemokine [C-C motif] receptor 7, fold change =-4.27, adjusted p<0.001) during the initial fatigue intensification (Table 3). In a parallel ongoing animal model of fatigue-induced by radiation, similar gene expression patterns were found (Table 4).
| Probeset ID | Gene Symbol | Fold | p-value | Gene Title |
|---|---|---|---|---|
| 206177_s_at | ARG1 | 2.41568 | 0.0000344 | arginase, liver |
| 1418847_at | Arg2 | 3.66503 | 4.01921E-06 | arginase type II |
| 205758_at | CD8A | -2.1743 | 3.76E-08 | CD8a antigen molecule |
| 1451673_at | Cd8a | -2.64612 | 2.63446E-05 | CD8 antigen, alpha chain |
| 1426170_a_at | Cd8b1 | -3.54196 | 1.92496E-06 | CD8 antigen, beta chain 1 |
| 206545_at | CD28 | -2.11077 | 6.71E-09 | CD28 molecule |
| 1437025_at | Cd28 | -2.78915 | 8.28414E-07 | CD28 antigen |
| 206150_at | CD27 | -2.10736 | 1.24E-14 | CD27 molecule |
| 1452389_at | Cd27 | -2.32832 | 8.33527E-06 | CD27 antigen |
| 206337_at | CCR7 | -4.26661 | 4.73E-14 | chemokine (C-C motif) receptor 7 |
| 1423466_at | Ccr7 | -6.26659 | 1.0981E-09 | chemokine (C-C motif) receptor 7 |
| 213539_at | CD3D | -2.05128 | 2.97753E-08 | CD3d molecule, delta (CD3-TCR complex) |
| 206804_at | CD3G | -2.09743 | 3.32498E-09 | CD3g molecule, gamma (CD3-TCR complex) |
| 1422828_at | Cd3d | -2.49422 | 2.25E-05 | CD3 antigen, delta polypeptide |
| 1419178_at | Cd3g | -4.40508 | 3.17E-07 | CD3 antigen, gamma polypeptide |
| 220059_at | STAP1 | -2.77349 | 2.74E-15 | signal transducing adaptor family member 1 |
| 1421098_at | Stap1 | -1.99621 | 0.00011027 | signal transducing adaptor family member 1 |
| 226218_at | IL7R | -2.87866 | 9.74E-10 | interleukin 7 receptor |
| 1448575_at | Il7r | -4.00297 | 4.4183E-08 | interleukin 7 receptor |
| 212827_at | IGHM | -6.84302 | 3.81E-19 | immunoglobulin heavy constant mu |
| 1427329_a_at | Igh-6 | -4.69937 | 8.32855E-08 | immunoglobulin heavy chain 6 |
| 1455530_at | Ighv14-2 | -16.3121 | 3.64611E-11 | immunoglobulin heavy variable V14-2 |
| 1421653_a_at | Igh-2 | -10.2349 | 8.58544E-06 | immunoglobulin heavy chain 2 (serum IgA) |
| 231124_x_at | LY9 | -2.72628 | 1.4E-13 | lymphocyte antigen 9 |
| 1416930_at | Ly6d | -4.76003 | 5.51E-07 | lymphocyte antigen 6 complex, locus D |
| 227646_at | EBF1 | -2.61277 | 1.81E-16 | early B-cell factor 1 |
| 1416301_a_at | Ebf1 | -2.29925 | 4.28E-05 | early B-cell factor 1 |
Table 4: Comparison of changes in gene expression following EBRT in human and mouse.
▪ Verification of changes in gene expression by qRT-PCR
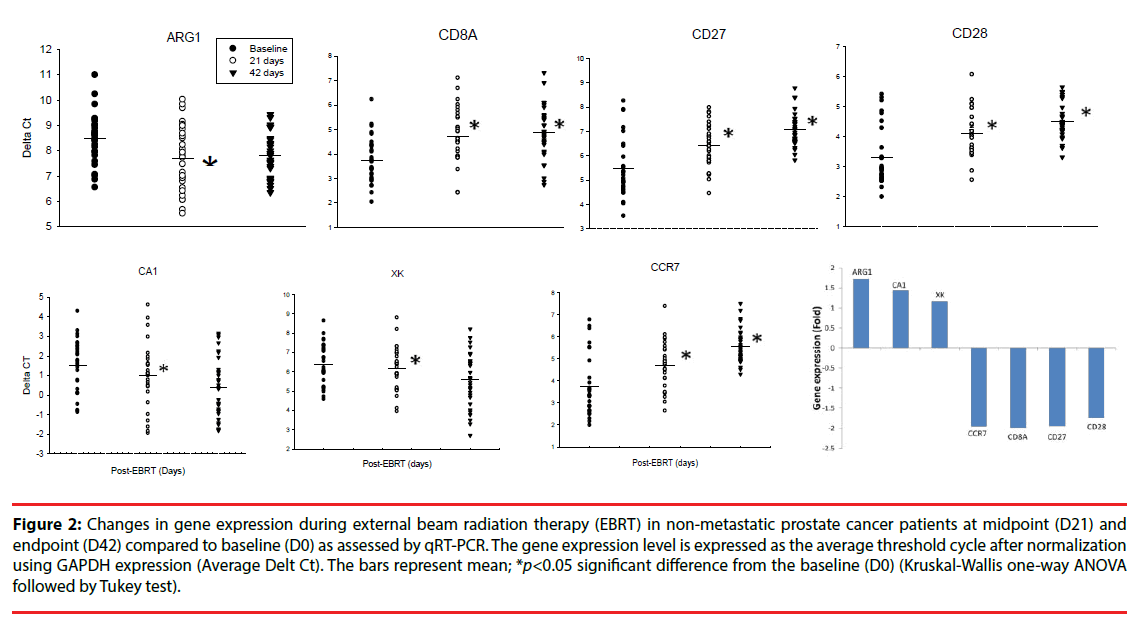
The changes in expression of the genes of interest selected from the microarray findings were further verified using qRT-PCR. ARG1 was significantly upregulated at D21 of EBRT compared to D0 (Figure 2); (1.72-fold, p=0.017, Kruskal-Wallis One Way Analysis of Variance on Ranks followed by post hoc Tukey - test). In contrast, the expressions of CD8A, CD27, CD28, and CCR7, were significantly downregulated (p<0.05) at D21 of EBRT (Figure 2), CD8A 1.98-fold, CD27 1.94-fold, CD28 1.74-fold, and CCR7 1.95-fold) compared to baseline.
Figure 2: Changes in gene expression during external beam radiation therapy (EBRT) in non-metastatic prostate cancer patients at midpoint (D21) and endpoint (D42) compared to baseline (D0) as assessed by qRT-PCR. The gene expression level is expressed as the average threshold cycle after normalization using GAPDH expression (Average Delt Ct). The bars represent mean; *p<0.05 significant difference from the baseline (D0) (Kruskal-Wallis one-way ANOVA followed by Tukey test).
▪ Correlation of the changes in the expression genes of interest related to fatigue and their association with fatigue intensity during EBRT
To further examine whether these genes of interest encoding T-cell signaling pathway have a similar expression profile and whether they participate in a common or intersecting biological pathway in the fatigue intensification during EBRT, the Pearson correlation coefficients were calculated among these genes. As shown in Table 5, the upregulation of ARG1 was highly correlated with the changes in the gene expressions of CCR7, CD27, CD28, CD8A, XK and CA1. Moreover, there were significant correlations among the changes in expression of the genes related to T-cell proliferation CCR7, CD27, CD28 and CD8A during EBRT.
| Gene Symbol | Arg1 | CCR7 | CD27 | CD28 | CD8A |
|---|---|---|---|---|---|
| Arg1 | 1 | 0.93577 (1.31E-41) | 0.960055 (1.86E-50) |
0.947467 (2.44E-45) |
0.93528 (1.82E-41) |
| CCR7 | 1 | 0.991251 (3.47E-79) |
0.994166 (6.65E-87) |
0.962445 (1.29E-51) | |
| CD27 | 1 | 0.993718 (1.71E-85) |
0.98082 (2.78E-64) |
||
| CD28 | 1 | 0.974993 (2.88E-59) | |||
| CD8A | 1 |
Table 5: Association among gene Arg1, CCR7, CD27, CD28, and CD8A expression (delta Ct) presented as Pearson correlation coefficients (p value).
More interestingly, the upregulation of ARG1 gene expressions following EBRT was positively correlated with the reported fatigue scores patients as measured by the PROMIS-F (higher score indicating higher level of fatigue) whereas the upregulation of the gene expression of CCR7, CD27 and CD28 was negatively correlated to the patient reported fatigue intensity (Table 6).
| Gene name | r value | p value |
|---|---|---|
| ARG1 | 0.2285 | 0.040192 |
| XK | 0.120924 | 0.282218 |
| CA1 | 0.227495 | 0.041102 |
| CCR7 | -0.28672 | 0.009456 |
| CD27 | -0.25271 | 0.022839 |
| CD28 | -0.26166 | 0.018292 |
| CD8A | -0.15419 | 0.169327 |
Table 6: Correlation between gene expression of interest and fatigue intensity (PROMIS) following RBRT.
▪ Correlation between ARG1 gene expression and lymphocyte count
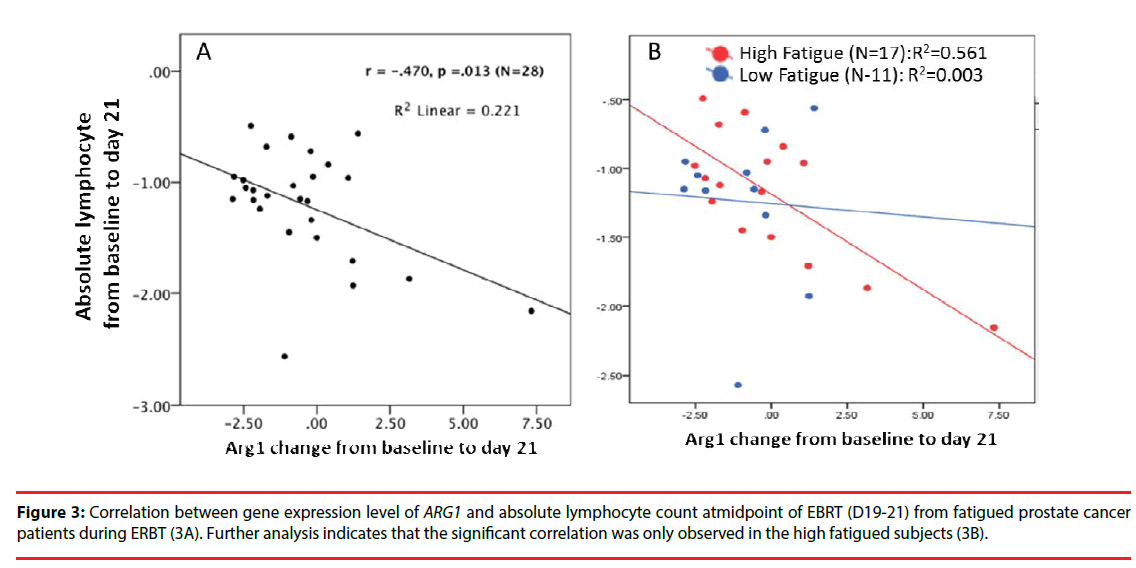
Since the pathway analysis indicated that these specific differentially expressed genes of interest are mainly associated with the T lymphocyte signaling pathway (p=3.48 × 1012) during EBRT, we further examined whether the upregulation of ARG1 is associated with the changes in lymphocyte account during EBRT. As shown in Figure 3, EBRT-related upregulation of ARG1 was negatively correlated with the change in absolute lymphocyte count (Figure 3A), r=- 0.47, p=0.01) during EBRT. Further analysis between the high versus low fatigue groups (high fatigue, >3-point change in FACT-F score from baseline to midpoint of EBRT), the association of ARG1 expression and change in absolute lymphocyte count was only observed in the high fatigue group (Figure 3B), r=-0.56) but not in the low fatigue group (r=-0.003).
▪ Differential expression levels of arginase I and arginine in plasma between the high and low fatigue subjects
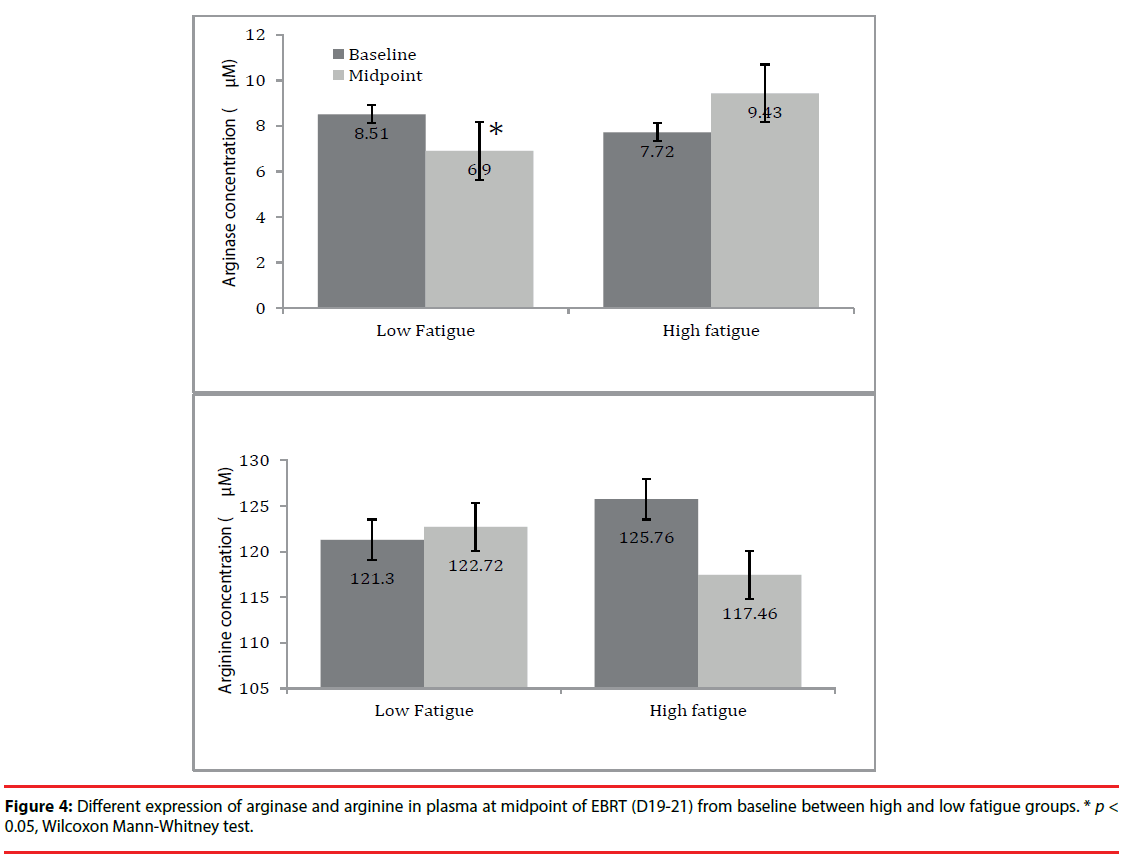
Finally, the arginase I and arginine levels in plasma were examined based on their metabolic pathway. As shown in (Figure 4), the arginase level in plasma showed an increased trend in high fatigue (HF) group at midpoint (9.4 ± 8.8) compared to baseline (7.7 ± 2.4); whereas, in the low fatigue (LF) group, arginase level in plasma significantly declined at midpoint (6.9 ± 2.4) from baseline (8.5 ± 4.6, p=0.02). For arginine, its level in plasma decreased at midpoint (117.5 ± 26.1) compared to baseline (125.8 ± 30.7) in the HF group and remained unchanged at midpoint (121.7 ± 28.1) from baseline (121.3 ± 26.8, p= 0.82) in the LF group. However, the changes in arginase and arginine in the HF group did not reach statistical significance, possibly due to the large inter-individual variation and limited sample size.
Discussion
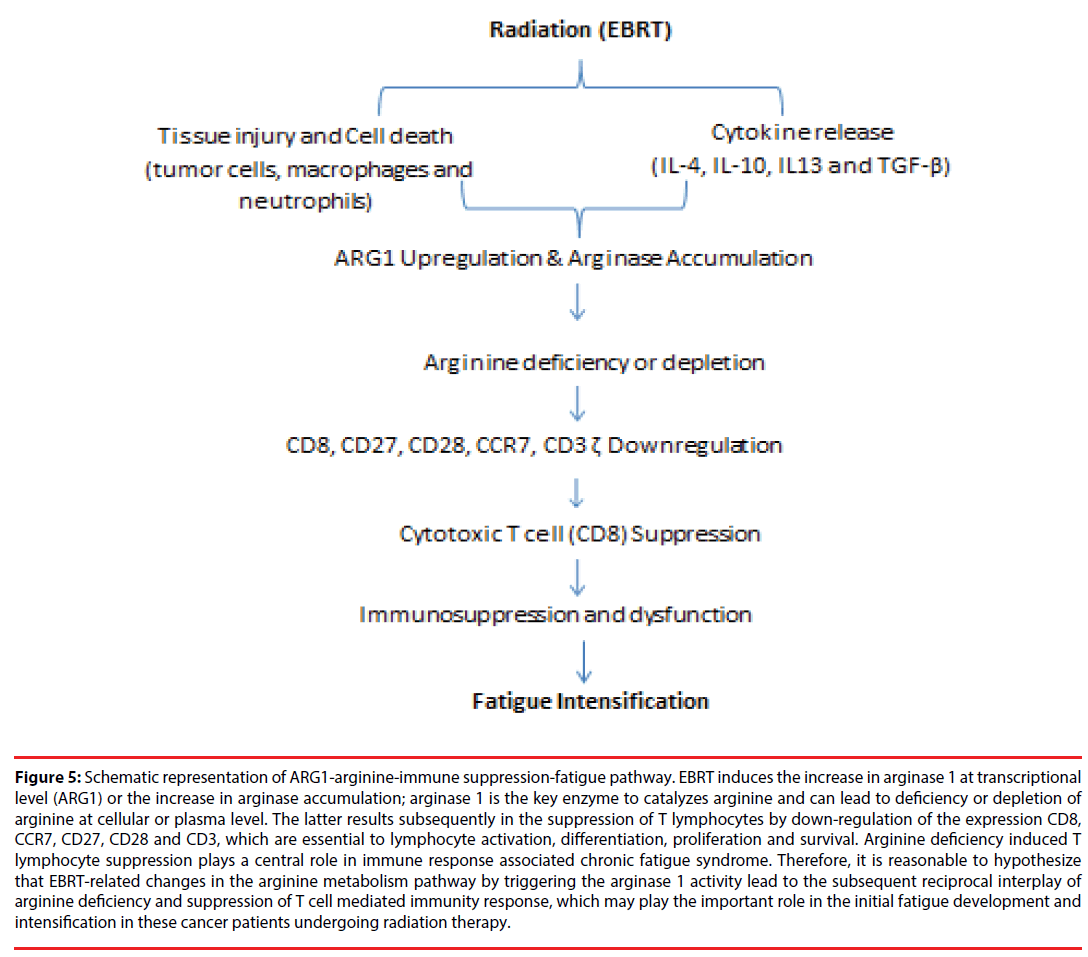
The principal novel finding from this study is the increase in the expression of arginase 1 ARG1 that is correlated with the decrease in the absolute lymphocyte count in the prostate cancer patients who developed significant fatigue during EBRT. Moreover, the highly significant correlation between expression of ARG1 and the expression of T-cell proliferation markers of CD8, CD28, CD27, and CCR7 indicate that ARG1 mRNA levels were mainly expressed by T lymphocytes. This latter finding has been previously reported [23] and these protein encoding genes are noted to be highly expressed in less-differentiated T cells [24,25]. Although the changes did not reach statistical significance, there was a trend for HF subjects to show an increase in arginase and a decrease in arginine in plasma at midpoint when compared to baseline. Our findings are consistent with previous reports that T lymphocytes are suppressed by depletion of L-arginine through increased expression and activity of arginase 1 [26]. Moreover, our findings indicate that the reciprocal interplay of arginine metabolism and T lymphocyte activity with fatigue development and intensification during localized RT in non-metastatic prostate cancer patients. We hypothesize that fatigue intensification during EBRT in this clinical population may be associated with: (1) EBRTrelated changes in the arginine metabolism pathway that trigger the arginase 1 activity, and (2) the subsequent reciprocal interplay of arginine deficiency and T cell receptor signaling downregulation causing suppression of T cell mediated immune response (Figure 5).
Figure 5: Schematic representation of ARG1-arginine-immune suppression-fatigue pathway. EBRT induces the increase in arginase 1 at transcriptional level (ARG1) or the increase in arginase accumulation; arginase 1 is the key enzyme to catalyzes arginine and can lead to deficiency or depletion of arginine at cellular or plasma level. The latter results subsequently in the suppression of T lymphocytes by down-regulation of the expression CD8, CCR7, CD27, CD28 and CD3, which are essential to lymphocyte activation, differentiation, proliferation and survival. Arginine deficiency induced T lymphocyte suppression plays a central role in immune response associated chronic fatigue syndrome. Therefore, it is reasonable to hypothesize that EBRT-related changes in the arginine metabolism pathway by triggering the arginase 1 activity lead to the subsequent reciprocal interplay of arginine deficiency and suppression of T cell mediated immunity response, which may play the important role in the initial fatigue development and intensification in these cancer patients undergoing radiation therapy.
▪ Immune dysfunction and fatigue
It is well known that adaptive immune response including T helper (TH) and cytotoxic T lymphocytes (CTL) occurs during the early stage of tumor growth and results in the development of CD8+ T cells reactive to tumors [27]. The CD8 antigen is found in all cytotoxic T lymphocytes (CD8+ T cells) and acts as the key player in antitumor immune response by antigen recognition. However, naïve CD8+ T cells need a co-stimulatory signal to activate and maintain their optimal activation [28]. CD27, CD28 and CCR7, which are highly expressed on naive CD8+ T cells [25], are essential to co-stimulate CD8+ T cell activation, proliferation, survival, and differentiation [29-31].
The CD8+T cells have consistently been shown to be decreased in patients with chronic fatigue [32-34]. One of the differences between patients with chronic fatigue and healthy controls was the striking down-regulation of CD+8 T cells with significantly lower activation, proliferative response and frequency of effector memory cells in individuals with chronic fatigue [24]. Furthermore, the CD27 receptor transduces signals that lead to the activation of NF-kappaB and MAPK/JNK pathways, which are also associated with the development of chronic fatigue pathways [35-38]. The current findings of decreased expression of CD8, CD28 CD27, CCR7 and CD3 during EBRT suggest that a CD+8 T-cell suppressed responses may play a key role in the initial development of fatigue in cancer patients undergoing radiation therapy (RT).
▪ Arginine deficiency, cell dysfunction, and immune suppression
One of the mechanisms underlying T lymphocyte cell-mediated fatigue may be related to the activities of L-arginine metabolism enzyme, arginase 1 [39]. Arginase remains a crucial player in host defense; specifically arginase 1, which is selectively expressed in granulocytes (polymorphonuclear leukocytes) where it modulates the immune system through arginine depletion [40]. It was reported that increased arginase 1 (ARG 1) activity in prostate cancer and lung cancer patients is involved in peripheral immunosuppression [41-43]. High arginase 1 activity depletes L-arginine and the downstream effects of which can inhibit CD8+ T-cells proliferation and activation, or trigger T-cell apoptosis because arginine is required for T-cell receptor formation and proliferation [44-46]. Activated CD8+ T-cells are particularly sensitive to the L-arginine level in their microenvironment [47]. Low or absent L-arginine significantly decreases proliferation of T-cells in vitro and activated T-cells are arrested in the G0-G1 phase of cell cycle; whereas T-cells cultured with L-arginine have their proliferation restored into the S and G2-M phases [48].
The main effect of over expression of arginase 1 on T-cell suppression is caused by L-arginine depletion [23,49] because L-arginine, a semiessential amino acid, plays a central role in immune response through the arginine metabolic pathway [43]. Arginine metabolism through arginase 1 produces urea and ornithine [50]. The latter is a precursor of different products, including polyamines and proline, which play a key role in cell proliferation [51] and wound healing [43]. T lymphocytes depend on arginine for multiple key biological processes, including proliferation, expression of the TCR complex and the ζ-chain peptide, and the development of memory T-cells [52]. With arginine deprivation, there is a progressive reduction (to ∼25% of basal levels) in the number of T-cell receptors on the cell membrane, as observed in certain cancers and after surgery or trauma [53,54]. A few hours after injury, a decrease in circulating arginine and an increase in accumulating arginase 1 expressed in immature myeloid cells further indicate that the arginine deficit is largely influenced by liberated arginase, rather than lack of arginine intake [52]. It is worth mentioning that the release of cytokines (IL-4, IL-10 and IL-13), catecholamines, and prostaglandin E2 (PGE2) by malignant tumors and adjacent tissue may also lead to arginase accumulation that enables synergism to deplete serum and cellular arginine, and ultimately lead to the depletion of arginine and result in impairment of T-cell proliferative response [55].
The current findings show the differential expression of ARG1 gene, increased arginase 1 level, and decreased arginine concentration in plasma of prostate cancer patients developed fatigue during EBRT. Furthermore, the decreased lymphocyte count in HF subjects during EBRT suggests the interplay between arginine metabolic pathway and CD8+ T-cell function as the potential mechanisms to explain fatigue intensification in these subjects. We hypothesize that the arginineimmune suppression-fatigue pathway shown in (Figure 5) triggered by EBRT may explain how EBRT causes an over expression of arginase, and how arginine depletion causes suppression of T-cell function and translates into a change in behavior, such as fatigue intensification.
▪ Clinical implications
Existing arginase inhibitors or exogenous arginine administration have been shown to re-establish T-cell proliferation [56,57]. While several caveats should be taken into consideration when evaluating the current findings, we discuss the potential strategies for clinical consideration to relieve the debilitating fatigue associated with cancer treatment.
Blockade of arginase expression and upregulation: Arginase inhibitors N-Hydroxy-nor-l-Arg (Nor- NOHA) and N-Hydroxy-l-Arg (NOHA) have been shown to partially prevent the reduction of l-arginine in culture T-cells and in an animal model [58]. Therefore, Arg-1 inhibitors may represent a target for development of new therapies to alleviate fatigue related to cancer and cancer treatment.
Arginine supplementations: increasing arginine level by exogenous L-arginine or dietary use of arginine supplementations have been shown to improve immune response in surgical patients. Citrulline, an arginine precursor, has been shown to counter high arginase activity [59].
Because tumor cells and PMNs release arginase through the COX-PGE E2 pathway [52,60], cyclooxygenase-2 (COX-2) inhibitors (e.g., celecoxib and sc-59125) could be used to reduce or prevent fatigue since these agents have been shown to prevent arginase 1 over expression and inhibit tumor growth [61].
Conclusions
Our current findings indicate that increased arginase 1 expression at the transcriptional level may play a critical role in fatigue intensification during EBRT, possibly through arginine deficiency-associated down-regulation on T-cell proliferation. These findings provide evidence at the transcriptional level of possible biologic correlates of cancer treatment-related fatigue, which may provide important insights into the molecular-genetic mechanisms that could be used in the development of new strategies for its prevention and treatment.
References
- Saligan LN, Kim HS. A systematic review of the association between immunogenomic markers and cancer-related fatigue. Brain. Behav. Immun 26(6), 830-848 (2012).
- Saligan LN, Hsiao CP, Wang D, et al. Upregulation of alpha-synuclein during localized radiation therapy signals the association of cancer-related fatigue with the activation of inflammatory and neuroprotective pathways. Brain. Behav. Immun 27(1), 63-70 (2013).
- Berger AM, Abernethy AP, Atkinson A, et al. NCCN Clinical Practice Guidelines Cancer-related fatigue. J. Natl. Compr. Canc. Netw 8(8), 904-931 (2010).
- Wang XM, Walitt B, Saligan L, et al. Chemobrain: a critical review and causal hypothesis of link between cytokines and epigenetic reprogramming associated with chemotherapy. Cytokine 72(1), 86-96 (2015).
- Prue G, Rankin J, Allen J, Gracey J, Cramp F, Cancer-related fatigue, A critical appraisal. Eur. J. Cancer 42(7),846-863 (2006).
- Henry DH, Viswanathan HN, Elkin EP, et al. Symptoms and treatment burden associated with cancer treatment, results from a cross-sectional national survey in the U.S. Support. Care. Cancer 16(7), 791-801 (2008).
- Servaes P, Gielissen MF, Verhagen S, et al. The course of severe fatigue in disease-free breast cancer patients, a longitudinal study. Psychooncology 16(9), 787-795 (2007).
- Galiano-Castillo N, Ariza-Garcia A, Cantarero-Villanueva I, et al. Depressed mood in breast cancer survivors, associations with physical activity, cancer-related fatigue, quality of life, and fitness level. Eur. J. Oncol. Nurs 18(2), 206-210 (2014).
- Fransson P. Fatigue in prostate cancer patients treated with external beam radiotherapy, a prospective 5-year long-term patient-reported evaluation. J. Cancer. Res. Ther 6(4), 516-520 (2010).
- Wong WW, Vora SA, Schild SE, et al. Radiation dose escalation for localized prostate cancer, intensity-modulated radiotherapy versus permanent transperineal brachytherapy. Cancer 115(23), 5596-5606 (2009).
- Budaus L, Bolla M, Bossi A, et al. Functional outcomes and complications following radiation therapy for prostate cancer, a critical analysis of the literature. Eur. Urol 61(1), 112-127 (2012).
- Langston B, Armes J, Levy A, et al. The prevalence and severity of fatigue in men with prostate cancer, a systematic review of the literature. Support. Care. Cancer 21(6), 1761-1771 (2013).
- NCCN, National Comprehensive Cancer Network, NCCN Clinical Practice Guidelines in Oncology, Cancer-Related Fatigue In. Edited by Fort Washington PNCCN; (2014).
- National Comprehensive Cancer Network, NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines), Cancer-Related Fatigue,(2016).
- Qu D, Zhang Z, Yu X, et al. Psychotropic drugs for the management of cancer-related fatigue, a systematic review and meta-analysis. Eur. J. Cancer. Care 25(6), 970-979 (2015).
- Bower JE, Ganz PA, Aziz N, et al. T-cell homeostasis in breast cancer survivors with persistent fatigue. J. Natl. Cancer. Inst 95(15), 1165-1168 (2003).
- Collado-Hidalgo A, Bower JE, Ganz PA, et al. Inflammatory biomarkers for persistent fatigue in breast cancer survivors. Clin. Cancer. Res 12(9), 2759-2766 (2006).
- Cella D, Lai JS, Chang CH, et al. Fatigue in cancer patients compared with fatigue in the general United States population. Cancer 94(2), 528-538 (2002).
- Cella D, Eton DT, Lai JS, et al. Combining anchor and distribution-based methods to derive minimal clinically important differences on the Functional Assessment of Cancer Therapy (FACT) anemia and fatigue scales. J. Pain. Symptom. Manage 24(6), 547-561 (2002).
- Lydiatt WM, Denman D, McNeilly DP, et al. A randomized, placebo-controlled trial of citalopram for the prevention of major depression during treatment for head and neck cancer. Arch. Otolaryngol. Head. Neck. Surg 134(5), 528-535 (2008).
- Luneburg N, Xanthakis V, Schwedhelm E, et al. Reference intervals for plasma L-arginine and the L-arginine,asymmetric dimethylarginine ratio in the Framingham Offspring Cohort. J. Nutr 141(12), 2186-2190 (2011).
- Wang XM, Hamza M, Wu TX, et al. Upregulation of IL-6, IL-8 and CCL2 gene expression after acute inflammation, Correlation to clinical pain. Pain 142(3), 275-283 (2009).
- Heuvers ME, Muskens F, Bezemer K, et al. Arginase-1 mRNA expression correlates with myeloid-derived suppressor cell levels in peripheral blood of NSCLC patients. Lung. cancer 81(3), 468-474 (2013).
- Curriu M, Carrillo J, Massanella M, et al. Screening NK-, B- and T-cell phenotype and function in patients suffering from Chronic Fatigue Syndrome. J. Transl. Med 11(1), 68 (2013).
- Ishikawa T, Kokura S, Enoki T et al. Phase I clinical trial of fibronectin CH296-stimulated T cell therapy in patients with advanced cancer. PloS. one 9(1), e83786 (2014).
- Munder M. Arginase: an emerging key player in the mammalian immune system. Br. J. Pharmacol 158(3), 638-651 (2009).
- North RJ. Down-regulation of the antitumor immune response. Adv. Cancer. Res 45(1), 1-43 (1985).
- Lenschow DJ, Walunas TL, Bluestone JA. CD28/B7 system of T cell costimulation. Annu. Rev. Immunol 14(1), 233-258 (1996).
- Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat. Rev. Immunol 9(4), 271-285 (2009).
- Nolte MA, van Olffen RW, van Gisbergen KP, et al. Timing and tuning of CD27-CD70 interactions, the impact of signal strength in setting the balance between adaptive responses and immunopathology. Immunol. Rev 229(1), 216-231 (2009).
- Acuto O, Michel F, CD28-mediated co-stimulation, a quantitative support for TCR signalling. Nat. Rev. Immunol 3(12), 939-951 (2003).
- Brenu EW, Staines DR, Baskurt OK, et al. Immune and hemorheological changes in chronic fatigue syndrome. J. Transl. Med 8(1), 1 (2010).
- Klimas NG, Salvato FR, Morgan R, et al. Immunologic abnormalities in chronic fatigue syndrome. J. Clin. Microbiol 28(6), 1403-1410 (1990).
- Maher KJ, Klimas NG, Fletcher MA. Chronic fatigue syndrome is associated with diminished intracellular perforin. Clin. Exp. Immunol 142(3), 505-511(2005).
- Bower JE, Ganz PA, Irwin MR, et al. Fatigue and gene expression in human leukocytes, increased NF-kappaB and decreased glucocorticoid signaling in breast cancer survivors with persistent fatigue. Brain. Behav. Immun 25(1), 147-150 (2011).
- Morris G, Maes M. Increased nuclear factor-kappaB and loss of p53 are key mechanisms in Myalgic Encephalomyelitis/chronic fatigue syndrome (ME/CFS). Med. Hypotheses 79(5), 607-613 (2012).
- Powolny-Budnicka I, Riemann M, Tanzer S, et al. RelA and RelB transcription factors in distinct thymocyte populations control lymphotoxin-dependent interleukin-17 production in gammadelta T cells. Immunity 34(3), 364-374 (2011).
- Pritchard AL, Hayward NK. Molecular pathways, mitogen-activated protein kinase pathway mutations and drug resistance. Clin Cancer Res 19(9), 2301-2309 (2013).
- Gallina G, Dolcetti L, Serafini P, et al. Tumors induce a subset of inflammatory monocytes with immunosuppressive activity on CD8+ T cells. J. Clin. Invest 116(10), 2777-2790 (2006).
- Munder M, Mollinedo F, Calafat J, et al. Arginase I is constitutively expressed in human granulocytes and participates in fungicidal activity. Blood 105(6), 2549-2556 (2005).
- Bronte V, Kasic T, Gri G, et al. Boosting antitumor responses of T lymphocytes infiltrating human prostate cancers. J. Exp. Med 201(8), 1257-1268 (2005).
- Sippel TR, White J, Nag K, et al. Neutrophil degranulation and immunosuppression in patients with GBM, restoration of cellular immune function by targeting arginase I. Clin. Cancer Res 17(22), 6992-7002 (2011).
- Bronte V, Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat. rev. Immunol 5(8), 641-654 (2005).
- Condamine T, Gabrilovich DI. Molecular mechanisms regulating myeloid-derived suppressor cell differentiation and function. Trends. Immunol 32(1), 19-25 (2011).
- Dolcetti L, Marigo I, Mantelli B, et al. Myeloid-derived suppressor cell role in tumor-related inflammation. Cancer. let 267(2), 216-225 (2008).
- Rotondo R, Bertolotto M, Barisione G, et al. Exocytosis of azurophil and arginase 1-containing granules by activated polymorphonuclear neutrophils is required to inhibit T lymphocyte proliferation. J. Leukoc. Biol 89(5), 721-727 (2011).
- Frey AB. Myeloid suppressor cells regulate the adaptive immune response to cancer. J. Clin. Invest 116(10), 2587-2590 (2006).
- Rodriguez PC, Quiceno DG, Ochoa AC. L-arginine availability regulates T-lymphocyte cell-cycle progression. Blood 109(4), 1568-1573 (2007).
- Popovic PJ, Zeh HJ, Ochoa JB. Arginine and immunity. J Nutr 137(6), 1681S-1686S (2007).
- Morris SM Jr. Enzymes of arginine metabolism. J. Nutr 134(10), 2743S-2747S (2004).
- Bronte V, Serafini P, Mazzoni A, et al. L-arginine metabolism in myeloid cells controls T-lymphocyte functions. Trends. Immunol 24(6), 302-306 (2003.
- Ochoa JB, Strange J, Kearney P, et al. Effects of L-arginine on the proliferation of T lymphocyte subpopulations. 25(1), JPEN. J. Parenter. Enteral. Nutr 23-29 (2001).
- Kusmartsev S, Gabrilovich DI. Role of immature myeloid cells in mechanisms of immune evasion in cancer. Cancer. Immunol. Immunother 55(3), 237-245 (2006).
- Taheri F, Ochoa JB, Faghiri Z, et al. L-Arginine regulates the expression of the T-cell receptor zeta chain (CD3zeta) in Jurkat cells. Clin. Cancer. Res 7(3), 958-965 (2001).
- Raber P, Ochoa AC, Rodriguez PC. Metabolism of L-arginine by myeloid-derived suppressor cells in cancer, mechanisms of T cell suppression and therapeutic perspectives. Immunol. Invest 41(6-7), 614-634 (2012).
- Rodriguez E, Seguer J, Rocabayera X, et al. Cellular effects of monohydrochloride of L-arginine, N-lauroyl ethylester (LAE) on exposure to Salmonella typhimurium and Staphylococcus aureus. J. Appl. Microbiol 96(5), 903-912 (2004).
- Rodriguez PC, Ochoa AC. Arginine regulation by myeloid derived suppressor cells and tolerance in cancer, mechanisms and therapeutic perspectives. Immunol. Rev 222, 180-191 (2008).
- Makarenkova VP, Bansal V, Matta BM, et al. CD11b+/Gr-1+ myeloid suppressor cells cause T cell dysfunction after traumatic stress. J. Immunol 176(4), 2085-2094 (2006).
- Bansal V, Rodriguez P, Wu G, et al. Citrulline can preserve proliferation and prevent the loss of CD3 zeta chain under conditions of low arginine. JPEN. J. Parenter. Enteral. Nutr 28(6), 423-430 (2004).
- Zhu W, Chandrasekharan UM, Bandyopadhyay S, et al. Thrombin induces endothelial arginase through AP-1 activation. Am. J. Physiol. Cell. Physiol 298(4), C952-960 (2010).
- Rodriguez PC, Hernandez CP, Quiceno D, et al. Arginase I in myeloid suppressor cells is induced by COX-2 in lung carcinoma. J. Exp. Med 202(7), 931-939 (2005).